
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,224,911 – 6,225,063 |
| Length | 152 |
| Max. P | 0.823146 |

| Location | 6,224,911 – 6,225,001 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -41.72 |
| Consensus MFE | -34.72 |
| Energy contribution | -35.34 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6224911 90 + 27905053 UUGGCAAGCAACAGCAACAACAGGCUGUGGUGGCCGCCGG---AGCGGGACUUCCGCUGUCCACCAACUCGGCGGCGGCUCAGGCAGCCGCUG ..(((..((....)).......(((((((((.((((((((---(((((.....)))))((......)))))))))).)))...))))))))). ( -42.30) >DroSec_CAF1 42125 90 + 1 UUGGCAAGCAACAGCAACAACAGGCUGUGGUGGCCGCCGG---AGCGGGACUUCCGCUGUCCACCAACUCGGCGGCGGCUCAGGCGGCCGCUG ..........(((((........)))))((((((((((.(---(((.((....))((((.((........)))))).)))).)))))))))). ( -45.40) >DroSim_CAF1 43862 90 + 1 UUGGCAAGCAACAGCAACAACAGGCUGUGGUGGCCGCCGG---AGCGGGACAUCCGCUGUCCACCAACUCGGCGGCGGCUCAGGCGGCCGCUG ..........(((((........)))))((((((((((.(---(((.((....))((((.((........)))))).)))).)))))))))). ( -45.40) >DroEre_CAF1 42454 90 + 1 UUGGCAAGCAACAGCAACAGCAAGCUGUGGUGGCCGCCGG---AGCGGGACAUCCCCUGUCCACCAACUCGGCGGCGGCUCAGGCAGCCGCUG .......((....))..((((..((((((((.((((((((---.(..(((((.....)))))..)...)))))))).)))...))))).)))) ( -40.10) >DroYak_CAF1 39498 90 + 1 UUGGCAAGCAACAGCAACAACAGGCUGUGGUGGCCGCCGG---AGCGGGACCUCCCCUGUCCACCAACUCGGCGGCGGCUCAGGCAGCCGCUG ..(((..((....)).......(((((((((.((((((((---.(..((((.......))))..)...)))))))).)))...))))))))). ( -40.90) >DroAna_CAF1 47287 84 + 1 CCGGAAAGCAA---------CAGGCGGUGGUGGCCGCUGGAGGAGCUGGUGUCCCACUCUCCACGAGUUCGGCCGCUGCCCAGGCAGCCGCAG .(((...((..---------..((((((((((((((.(((((.((.(((....)))))))))))).)))..)))))))))...))..)))... ( -36.20) >consensus UUGGCAAGCAACAGCAACAACAGGCUGUGGUGGCCGCCGG___AGCGGGACUUCCGCUGUCCACCAACUCGGCGGCGGCUCAGGCAGCCGCUG ..(((..((....)).......(((((((((.((((((((.......((((.......))))......)))))))).)))...))))))))). (-34.72 = -35.34 + 0.61)



| Location | 6,224,971 – 6,225,063 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -33.40 |
| Energy contribution | -34.52 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6224971 92 + 27905053 CCAACUCGGCGGCGGCUCAGGCAGCCGCUGUGGCCGCCGCAGCGGUGGCUAAGGCCAAACUGGAGCAGGCCAUGAACCAGAGCUGGUAAAGC (((.(((((((((.((....)).))))))((((((((..(((...((((....))))..)))..)).))))))......))).)))...... ( -43.30) >DroVir_CAF1 42017 86 + 1 CUAGCUCAGCAGCCGUGCAGGCGGCUGCUGCUG------CCGCUGUGGCUAAGGCCAAGCUGGAGCAGGCCAUUAGCAACAGCUGGUAAGCU ..((((((((((((((....)))))))))).((------(((((((.(((((((((..((....)).)))).))))).))))).)))))))) ( -48.80) >DroSec_CAF1 42185 92 + 1 CCAACUCGGCGGCGGCUCAGGCGGCCGCUGUGGCCGCCGCAGCGGUGGCCAAGGCCAAACUGGAGCAGGCCAUGAACCAGAGCUGGUAAAGC ...((.((((...((((..(((.((((((((((...)))))))))).)))..))))...((((..((.....))..)))).))))))..... ( -47.20) >DroSim_CAF1 43922 92 + 1 CCAACUCGGCGGCGGCUCAGGCGGCCGCUGUGGCCGCCGCAGCGGUGGCCAAGGCCAAACUGGAGCAGGCCAUGAACCAGAGCUGGUAAAGC ...((.((((...((((..(((.((((((((((...)))))))))).)))..))))...((((..((.....))..)))).))))))..... ( -47.20) >DroYak_CAF1 39558 92 + 1 CCAACUCGGCGGCGGCUCAGGCAGCCGCUGUGGCCGCCGCUGCGGUGGCCAAGGCCAAACUGGAGCAGGCCAUGAACCAGAGCUGGUAAGGC (((.(((((((((.((....)).))))))((((((...(((.(((((((....)))..)))).))).))))))......))).)))...... ( -46.20) >DroAna_CAF1 47341 92 + 1 CGAGUUCGGCCGCUGCCCAGGCAGCCGCAGCAGCCGCCGCCGCAGUGGCUAAGGCCAAACUAGAGCAGGCCAUCAACCAGGGAUGGUGAAGU ...(((((((((((((...(((.((.((....)).)).)))))))))))).((......)).))))..((((((.......))))))..... ( -38.00) >consensus CCAACUCGGCGGCGGCUCAGGCAGCCGCUGUGGCCGCCGCAGCGGUGGCCAAGGCCAAACUGGAGCAGGCCAUGAACCAGAGCUGGUAAAGC ......(((((((.((....)).)))))))((((((((.....)))))))).((((...........))))....((((....))))..... (-33.40 = -34.52 + 1.11)

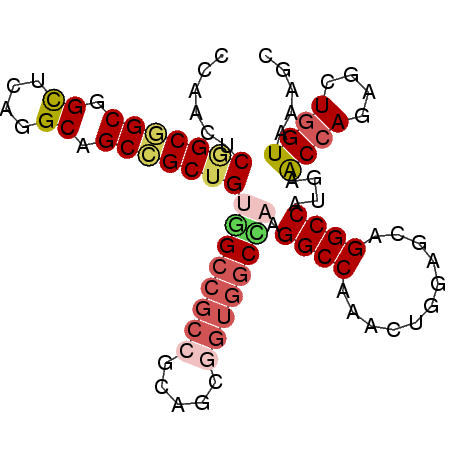

| Location | 6,224,971 – 6,225,063 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 83.77 |
| Mean single sequence MFE | -45.77 |
| Consensus MFE | -32.46 |
| Energy contribution | -33.88 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6224971 92 - 27905053 GCUUUACCAGCUCUGGUUCAUGGCCUGCUCCAGUUUGGCCUUAGCCACCGCUGCGGCGGCCACAGCGGCUGCCUGAGCCGCCGCCGAGUUGG ......(((((((.(((...(((((.(((.((((.((((....))))..)))).))))))))..((((((.....)))))).)))))))))) ( -46.50) >DroVir_CAF1 42017 86 - 1 AGCUUACCAGCUGUUGCUAAUGGCCUGCUCCAGCUUGGCCUUAGCCACAGCGG------CAGCAGCAGCCGCCUGCACGGCUGCUGAGCUAG ((((..((.(((((.(((((.((((.((....))..))))))))).)))))))------...(((((((((......))))))))))))).. ( -46.90) >DroSec_CAF1 42185 92 - 1 GCUUUACCAGCUCUGGUUCAUGGCCUGCUCCAGUUUGGCCUUGGCCACCGCUGCGGCGGCCACAGCGGCCGCCUGAGCCGCCGCCGAGUUGG ......(((((((.(((....(((..((((((((.((((....))))..)))).(((((((.....))))))).)))).))))))))))))) ( -48.70) >DroSim_CAF1 43922 92 - 1 GCUUUACCAGCUCUGGUUCAUGGCCUGCUCCAGUUUGGCCUUGGCCACCGCUGCGGCGGCCACAGCGGCCGCCUGAGCCGCCGCCGAGUUGG ......(((((((.(((....(((..((((((((.((((....))))..)))).(((((((.....))))))).)))).))))))))))))) ( -48.70) >DroYak_CAF1 39558 92 - 1 GCCUUACCAGCUCUGGUUCAUGGCCUGCUCCAGUUUGGCCUUGGCCACCGCAGCGGCGGCCACAGCGGCUGCCUGAGCCGCCGCCGAGUUGG ......(((((((.(((....((((.((....))..)))).(((((.(((...))).)))))..((((((.....)))))).)))))))))) ( -44.00) >DroAna_CAF1 47341 92 - 1 ACUUCACCAUCCCUGGUUGAUGGCCUGCUCUAGUUUGGCCUUAGCCACUGCGGCGGCGGCUGCUGCGGCUGCCUGGGCAGCGGCCGAACUCG ...((((((....))).)))((((((((((..((.((((....))))..))((((((.((....)).)))))).)))))..)))))...... ( -39.80) >consensus GCUUUACCAGCUCUGGUUCAUGGCCUGCUCCAGUUUGGCCUUAGCCACCGCUGCGGCGGCCACAGCGGCCGCCUGAGCCGCCGCCGAGUUGG ......((((((((((((...((((.((....))..))))..)))))..((.(((((((((.....)))))))...)).))....))))))) (-32.46 = -33.88 + 1.42)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:29:01 2006