
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
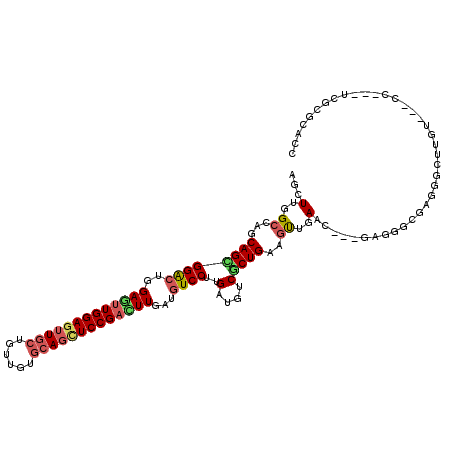
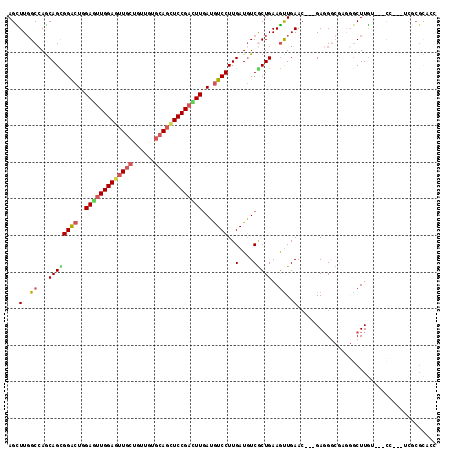
| Location | 6,224,513 – 6,224,655 |
| Length | 142 |
| Max. P | 0.883695 |

| Location | 6,224,513 – 6,224,615 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 79.26 |
| Mean single sequence MFE | -42.80 |
| Consensus MFE | -26.47 |
| Energy contribution | -27.03 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6224513 102 - 27905053 AGUUUGGCCAGCAGUGGGCUGGAGUUGGAGUUACCGUUGUGCAGCUCCGCCUUGAUGUCCUUGAUGUCGCUGAAGUUGAAC---GAGGGCGAGGGCUUGU---CC---------AGC .(((..((((((...((((.(((((((.(..........).))))))))))).((((((...))))))))))..))..)))---..((((((....))))---))---------... ( -38.30) >DroPse_CAF1 47671 111 - 1 AGCUUGGCCAGCAGCGGACUGGAGUUGGAGUUGCUGUUGUGCAGCUCCGACUUGAUGUCCUUGAUGUCGCUGAAGUUGAAC---GAGGGCGAGGGCUUGCUGUCC---UCGCGCAGC .(.(..((...(((((((((.(((((((((((((......))))))))))))).).))))..(....)))))..))..).)---..(.((((((((.....))))---)))).)... ( -55.20) >DroGri_CAF1 44960 105 - 1 AGCUUUGCGAGCAGUGGACUGGAGUUGGAGCUGCUGUUGUGCAAUUCCGAUUUGAUGUCCUUGAUGUCACUGAAGUUGAAC---GAGG------GCUUGU---UGUCUUCGCGCCCC (((((((.((.(((.(((((.(((((((((.(((......))).))))))))).).)))))))...)).).))))))...(---((((------((....---.)))))))...... ( -34.80) >DroWil_CAF1 57337 114 - 1 AGUUUGGCCAACAGUGGACUGGAAUUGGAGUUGCUGUUGUGCAGCUCCGAUUUAAUAUCCUUGAUGUCGCUGAAAUUGAACGUCGACGGCGAGGGCUUAU---CCUCGUUGCGUACC ..((..((..(((..(((...(((((((((((((......)))))))))))))....)))....))).))..)).......((((.(((((((((....)---)))))))))).)). ( -43.90) >DroMoj_CAF1 44327 102 - 1 AGCUUGGCCAGCAGCGGACUAGAAUUGGAAUUGGUAUUGUGCAGCUCCGACUUGAUGUCCUUGAUAUCACUGAAGCUGAAG---GAGG------GCUUGU---CG---UCACGCCCC .......((..((((((((..((.(((((.(((........))).))))).))...)))).((....)).....))))..)---).((------((.((.---..---.)).)))). ( -28.80) >DroPer_CAF1 47992 111 - 1 AGCUUGGCCAGCAGCGGACUGGAGUUGGAGUUGCUGUUGUGCAGCUCCGACUUGAUGUCCUUGAUGUCGCUGAAGUUGAAC---GAGGGCGAGGACUUGGUGUCC---UCGCGCAGC .(.(..((...(((((((((.(((((((((((((......))))))))))))).).))))..(....)))))..))..).)---..(.((((((((.....))))---)))).)... ( -55.80) >consensus AGCUUGGCCAGCAGCGGACUGGAGUUGGAGUUGCUGUUGUGCAGCUCCGACUUGAUGUCCUUGAUGUCGCUGAAGUUGAAC___GAGGGCGAGGGCUUGU___CC___UCGCGCACC ...(..((...((((((((..(((((((((((((......)))))))))))))...))))..(....)))))..))..)...................................... (-26.47 = -27.03 + 0.56)

| Location | 6,224,535 – 6,224,655 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -26.50 |
| Energy contribution | -27.95 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.865759 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
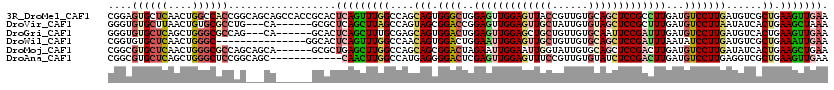
>3R_DroMel_CAF1 6224535 120 - 27905053 CGGAGUGCUCAACUGGCCACCGGCAGCAGCCACCGCACUCAGUUUGGCCAGCAGUGGGCUGGAGUUGGAGUUACCGUUGUGCAGCUCCGCCUUGAUGUCCUUGAUGUCGCUGAAGUUGAA ..((((((......((...))(((....)))...))))))..((..((((((...((((.(((((((.(..........).))))))))))).((((((...))))))))))..))..)) ( -41.10) >DroVir_CAF1 41605 111 - 1 GGGUGUGCUUAACUGUGCGCCUG---CA------GCGCUCAGCUUAGCCAGUAGCGGACCGGAGUUGGAGUUGCUAUUGUGUAGCUCCGCUUUGAUGUCCUUAAUAUCACUGAAGCUAAA (((((..(......)..))))).---.(------((..((((....((.....))((((((((((.((((((((......))))))))))))))..)))).........)))).)))... ( -42.20) >DroGri_CAF1 44985 111 - 1 GGGUGUGCUCAGCUGGGCGCCAG---CA------GCACUCAGCUUUGCGAGCAGUGGACUGGAGUUGGAGCUGCUGUUGUGCAAUUCCGAUUUGAUGUCCUUGAUGUCACUGAAGUUGAA ....(((((..(((((...))))---))------))))(((((((((.((.(((.(((((.(((((((((.(((......))).))))))))).).)))))))...)).).)))))))). ( -43.60) >DroWil_CAF1 57371 105 - 1 CGGUGUGCUCAACUGGGC---------------GGCACUCAGUUUGGCCAACAGUGGACUGGAAUUGGAGUUGCUGUUGUGCAGCUCCGAUUUAAUAUCCUUGAUGUCGCUGAAAUUGAA ..((((((((....))))---------------.))))((((((((((..(((..(((...(((((((((((((......)))))))))))))....)))....))).))).))))))). ( -41.30) >DroMoj_CAF1 44349 114 - 1 CGGCGUGCUCAACUGGGCGCCAGCAGCA------GCGCUGAGCUUGGCCAGCAGCGGACUAGAAUUGGAAUUGGUAUUGUGCAGCUCCGACUUGAUGUCCUUGAUAUCACUGAAGCUGAA .(((((.(......).)))))..((((.------((((((.((...)))))).))((((..((.(((((.(((........))).))))).))...))))..............)))).. ( -39.10) >DroAna_CAF1 46920 108 - 1 CGGCGUGCUCAGCUGGGCUCCGGCAGC------------CAACUUGGCCAUGAGGGGACUCGAGUUGGAGUUUCCGUUGUGUAUCUCCGACUUGAUGUCCUUGAGGUCGCUGAAGUUGAA (((((..(((.(((((...))))).((------------(.....))).....(((((((((((((((((...(......)...))))))))))).)))))))))..)))))........ ( -44.80) >consensus CGGUGUGCUCAACUGGGCGCCAG___CA______GCACUCAGCUUGGCCAGCAGUGGACUGGAGUUGGAGUUGCUGUUGUGCAGCUCCGACUUGAUGUCCUUGAUGUCACUGAAGUUGAA ....((((((....))))))..................(((((((((....(((.((((..(((((((((((((......)))))))))))))...)))))))......)).))))))). (-26.50 = -27.95 + 1.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:58 2006