
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,177,826 – 6,178,037 |
| Length | 211 |
| Max. P | 0.946222 |

| Location | 6,177,826 – 6,177,945 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -21.82 |
| Energy contribution | -21.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
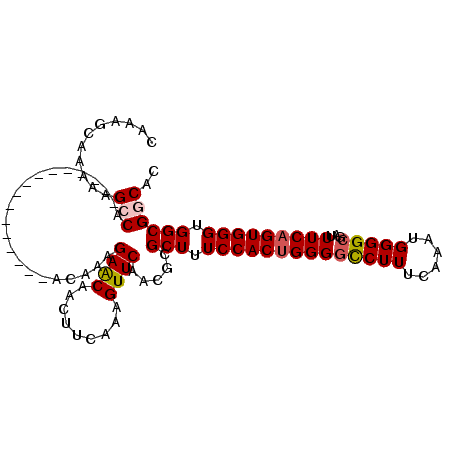
>3R_DroMel_CAF1 6177826 119 + 27905053 CAAAAGCUUUUCUCCCACCUUCUC-CAUCUCUUUCGCACUGUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAU ........................-...........(((((...((((((((..(((......)))..)).((((((((.........))))))))))))))...))))).......... ( -24.10) >DroSec_CAF1 30047 119 + 1 CAAAAGCUUUUCUCCCACCUUCUC-CAUCUCUUUCGCACGGUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGACAGCAAAGAAAGAAAUUAGUGAAACAAAAAU ........................-......((((((..((((.((((((((..(((......)))..)).(((((((((......)).))))))))))))))))).))))))....... ( -22.20) >DroSim_CAF1 33611 119 + 1 CAAAAGCUUUUCUCCCACCUUCUC-CAUCUCUUUCGCACGGUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAU ........................-......((((((..((((.((((((((..(((......)))..)).((((((((.........)))))))))))))))))).))))))....... ( -23.20) >DroEre_CAF1 29312 120 + 1 CAAAAGCUUUUCUCCCACCUUCGCCCAUCUCUUUCGCACCGUUCUCUUUCCUCUCCGAGGUUUCGGCAACCUUUGUUGCUGCAACAUGGCAGCAAAGAAAGAAAUUAGCGAAACAAAAAU ...................(((((..((.((((((...(((.((((..........))))...))).........(((((((......))))))).)))))).))..)))))........ ( -29.00) >DroYak_CAF1 30254 120 + 1 CAAAAGCUUUUCUCCCACCUUCUCCCAUCUCUUUCGCACUGUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCUGCAACAUAGCAGCAAAGAAAGAAAUUAGCGAAACAAAAAU ...............................((((((...(((.((((((((..(((......)))..)).....(((((((......))))))).)))))))))..))))))....... ( -28.00) >consensus CAAAAGCUUUUCUCCCACCUUCUC_CAUCUCUUUCGCACGGUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAU ...............................((((((...(((.((((((((..(((......)))..)).((((((((.........)))))))))))))))))..))))))....... (-21.82 = -21.98 + 0.16)


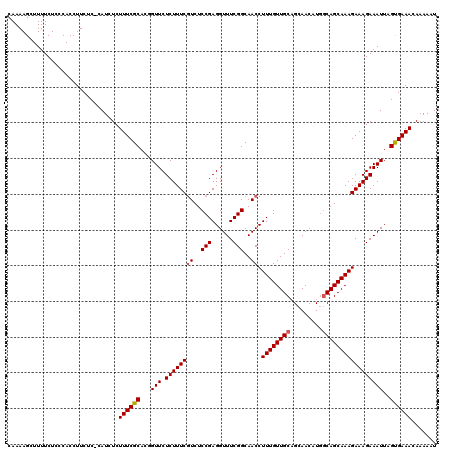
| Location | 6,177,826 – 6,177,945 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.49 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -30.00 |
| Energy contribution | -29.80 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6177826 119 - 27905053 AUUUUUGUUUCACUAAUUUCUUUCUUUGCUGCCAUGUUGCUGCAACAAAGGUUGCCGAAACCUCGGAGACGAAAGAGAACAGUGCGAAAGAGAUG-GAGAAGGUGGGAGAAAAGCUUUUG .(((((.(..((((..(((((.(((((..(((..((((.(((((((....))))).......(((....)))...))))))..)))..))))).)-)))).))))..).)))))...... ( -34.60) >DroSec_CAF1 30047 119 - 1 AUUUUUGUUUCACUAAUUUCUUUCUUUGCUGUCAUGUUGCUGCAACAAAGGUUGCCGAAACCUCGGAGACGAAAGAGAACCGUGCGAAAGAGAUG-GAGAAGGUGGGAGAAAAGCUUUUG .(((((.(..((((..((((((((.((((.((......)).)))).....((..((((....))))..)))))))))).((((.(....)..)))-)....))))..).)))))...... ( -34.10) >DroSim_CAF1 33611 119 - 1 AUUUUUGUUUCACUAAUUUCUUUCUUUGCUGCCAUGUUGCUGCAACAAAGGUUGCCGAAACCUCGGAGACGAAAGAGAACCGUGCGAAAGAGAUG-GAGAAGGUGGGAGAAAAGCUUUUG .(((((.(..((((..((((((((.((((.((......)).)))).....((..((((....))))..)))))))))).((((.(....)..)))-)....))))..).)))))...... ( -35.80) >DroEre_CAF1 29312 120 - 1 AUUUUUGUUUCGCUAAUUUCUUUCUUUGCUGCCAUGUUGCAGCAACAAAGGUUGCCGAAACCUCGGAGAGGAAAGAGAACGGUGCGAAAGAGAUGGGCGAAGGUGGGAGAAAAGCUUUUG ...(((.((((((...((((((((((((((((......)))))..(..(((((.....)))))..).))))))))))).....)))))).)))....(((((((.........))))))) ( -36.60) >DroYak_CAF1 30254 120 - 1 AUUUUUGUUUCGCUAAUUUCUUUCUUUGCUGCUAUGUUGCAGCAACAAAGGUUGCCGAAACCUCGGAGACGAAAGAGAACAGUGCGAAAGAGAUGGGAGAAGGUGGGAGAAAAGCUUUUG .(((((.(..((((..((((((((.(((((((......))))))).....((..((((....))))..)))))))))).((...(....)...))......))))..).)))))...... ( -38.50) >consensus AUUUUUGUUUCACUAAUUUCUUUCUUUGCUGCCAUGUUGCUGCAACAAAGGUUGCCGAAACCUCGGAGACGAAAGAGAACAGUGCGAAAGAGAUG_GAGAAGGUGGGAGAAAAGCUUUUG .(((((.(..((((..((((((((.((((.((......)).)))).....((..((((....))))..))))))))))......(....)...........))))..).)))))...... (-30.00 = -29.80 + -0.20)

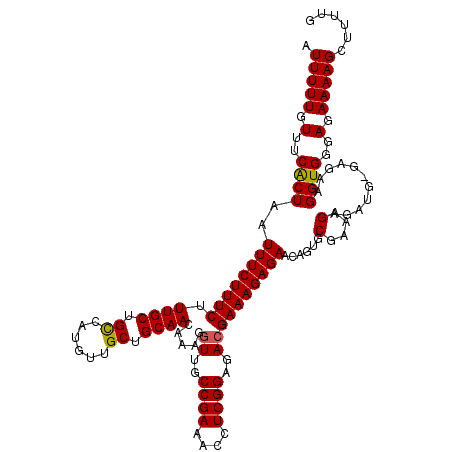
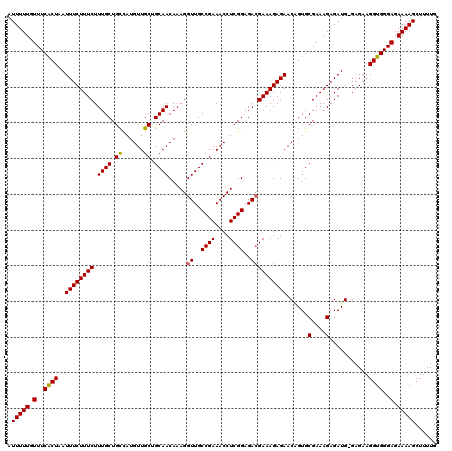
| Location | 6,177,865 – 6,177,969 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.31 |
| Mean single sequence MFE | -25.11 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.42 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6177865 104 + 27905053 GUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAUCAAAGCAAAAAGCCA----------------ACAAAGAAC (((((...(((....)))(((((..((...(((((((((.........)))))))))...........(((........)))..))...))))).----------------....))))) ( -22.30) >DroSec_CAF1 30086 104 + 1 GUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGACAGCAAAGAAAGAAAUUAGUGAAACAAAAAUCAAAGCAAAAAGCCA----------------ACAAAGAGC ...((((((((....)))(((((..((...((((((((((......)).))))))))...........(((........)))..))...))))).----------------...))))). ( -21.70) >DroSim_CAF1 33650 104 + 1 GUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAUCAAAGCAAAAAGCCA----------------ACAAAGAGC ...((((((((....)))(((((..((...(((((((((.........)))))))))...........(((........)))..))...))))).----------------...))))). ( -22.70) >DroEre_CAF1 29352 118 + 1 GUUCUCUUUCCUCUCCGAGGUUUCGGCAACCUUUGUUGCUGCAACAUGGCAGCAAAGAAAGAAAUUAGCGAAACAAAAAUCAAAUCAAAAAGCCA--GCCAGCAGAGCCCGGCAAAGAAC ((((((((((......((((((.....))))))..(((((((......))))))).)))))).............................(((.--(((....).))..)))...)))) ( -28.20) >DroYak_CAF1 30294 120 + 1 GUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCUGCAACAUAGCAGCAAAGAAAGAAAUUAGCGAAACAAAAAUCAAAGCAAAAAGCCAAAGCCAACAGAGCCCAGCAAAGAAC (((((...(((....)))(((((.(((...(((((((((((.....)))))))))))..........((...............)).....))).)))))...)))))............ ( -30.66) >consensus GUUCUCUUUCGUCUCCGAGGUUUCGGCAACCUUUGUUGCAGCAACAUGGCAGCAAAGAAAGAAAUUAGUGAAACAAAAAUCAAAGCAAAAAGCCA________________ACAAAGAAC (((((..((((....))))((((((....)(((((((((.........)))))))))............))))).........................................))))) (-19.26 = -19.42 + 0.16)


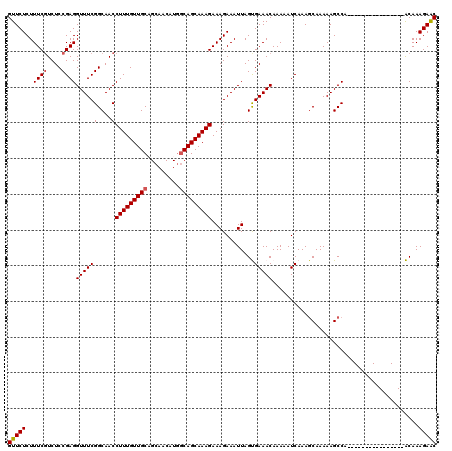
| Location | 6,177,945 – 6,178,037 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -25.50 |
| Energy contribution | -25.90 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6177945 92 + 27905053 CAAAGCAAAAAGCCA----------------ACAAAGAACAACUUCAAAGUUCAACGCGCUUUCCACUGGGGCCUUUCAAAUGGGGCGAUUUCAGUGGGUGGCGGCAC ...........(((.----------------.....((((.........)))).....(((.((((((((((((((......)))))...))))))))).)))))).. ( -30.60) >DroSec_CAF1 30166 92 + 1 CAAAGCAAAAAGCCA----------------ACAAAGAGCAACUUCAAAGUUCAACGCGCUUUCCACUGGGGCCUUUCAAAAGGGGCGAUUUCAGUGGGUGGCGACAC ....((.....))..----------------.....((((.........))))...(((((.((((((((((((((......)))))...))))))))).)))).).. ( -26.70) >DroSim_CAF1 33730 92 + 1 CAAAGCAAAAAGCCA----------------ACAAAGAGCAACUUCAAAGUUCAACGCGCUUUCCACUGGGGCCUUUCAAAUGGGGCGAUUUCAGUGGGUGGCGACAC ....((.....))..----------------.....((((.........))))...(((((.((((((((((((((......)))))...))))))))).)))).).. ( -26.90) >DroEre_CAF1 29432 106 + 1 CAAAUCAAAAAGCCA--GCCAGCAGAGCCCGGCAAAGAACAACUUCAAAGUUCAACGCGCUUUCCACUGGGCUCUUUCAAGUGGGGCGAUUUCAGUGGGUGGCGGCGC ...........(((.--((((..(((((.((.....((((.........))))..)).)))))((((((((((((.......)))))....))))))).))))))).. ( -33.70) >DroYak_CAF1 30374 108 + 1 CAAAGCAAAAAGCCAAAGCCAACAGAGCCCAGCAAAGAACAACUUCAAAGUUCAACGCGCUUUCCACUGGGGCCUUUCAAGUGGGGCGAUUUCUGUGGGUGGCGGCAC ...........(((...(((.(((((((((((....(((....)))(((((.......)))))...)))))(((((......)))))...)))))).)))...))).. ( -32.00) >consensus CAAAGCAAAAAGCCA________________ACAAAGAACAACUUCAAAGUUCAACGCGCUUUCCACUGGGGCCUUUCAAAUGGGGCGAUUUCAGUGGGUGGCGGCAC ...........(((......................((((.........)))).....(((.((((((((((((((......)))))...))))))))).)))))).. (-25.50 = -25.90 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:42 2006