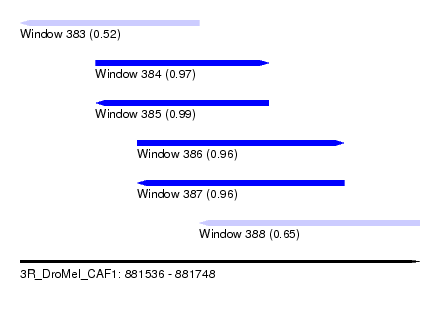
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 881,536 – 881,748 |
| Length | 212 |
| Max. P | 0.985418 |

| Location | 881,536 – 881,631 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -22.80 |
| Energy contribution | -23.93 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.69 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 881536 95 - 27905053 GGCCGUUCCCAGGCCUUCCUUCAUUCUGCUCGG-------------------------UCUGGUACACAAGCUCCGGGCUCAAAAUCAGGCCCGGCUCAUUCGUUAUGCGCGCACAAUGA ((((.......)))).....(((((.(((.(((-------------------------..((.....))..).(((((((........))))))).............)).))).))))) ( -31.00) >DroSec_CAF1 49577 95 - 1 GGCCUUUCCCAGGCCUUCCUUCAAUCUGCUCGG-------------------------UCUGGUCCACAAGCUCCGGGCUCAAAAUCAGGCCCGGCUCAUUCGUUAUGCGCGCACAAUGA (((((.....)))))............(((.((-------------------------((((((.....(((.....)))....)))))))).)))....(((((.((....)).))))) ( -28.20) >DroSim_CAF1 48780 95 - 1 GGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGG-------------------------UCUGGUCCACAAGCUCCGGGCUCAAAAUCAGGCCCGGCUCAUUCGUUAUGCGCGCACAAUGA (((((.....))))).....(((((.(((.(((-------------------------..((.....))..).(((((((........))))))).............)).))).))))) ( -31.30) >DroEre_CAF1 48538 116 - 1 GGCCAUUCCCAGGCCUUCG----UUCUGUUCGGUCUUCGUUCGGUCUGGUGUCUGUUGUCUGGUGCACAAGCUCCGGACUCGAAAUCAGGCCCGGCUCAUUCGCCAUGCACACACAAUGA ((((.......)))).(((----((.(((..(((((...((((((((((.(..(((.((.....)))))..).)))))).))))...))))).(((......)))......))).))))) ( -41.00) >consensus GGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGG_________________________UCUGGUCCACAAGCUCCGGGCUCAAAAUCAGGCCCGGCUCAUUCGUUAUGCGCGCACAAUGA ((((.......)))).....(((((.(((.((............................((.....)).(..(((((((........)))))))..)..........)).))).))))) (-22.80 = -23.93 + 1.13)

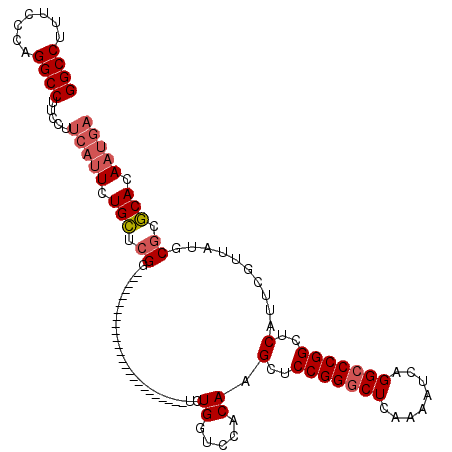
| Location | 881,576 – 881,668 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -40.90 |
| Energy contribution | -41.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.37 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 881576 92 + 27905053 AGCCCGGAGCUUGUGUACCAGACCGAGCAGAAUGAAGGAAGGCCUGGGAACGGCCGGGCAGACUGGUGGUCUGGAGCGGAGGGUCGGGCAUG .((((((..(((.(((.((((((((..(((..((......((((.(....)))))...))..))).)))))))).))).))).))))))... ( -45.90) >DroSec_CAF1 49617 92 + 1 AGCCCGGAGCUUGUGGACCAGACCGAGCAGAUUGAAGGAAGGCCUGGGAAAGGCCGUGCAGACUCGUGGUCUGGAGCGGAGGGUCGGGCAUG .((((((..(((.((..((((((((.(.((.(((......(((((.....)))))...))).))).))))))))..)).))).))))))... ( -41.80) >DroSim_CAF1 48820 92 + 1 AGCCCGGAGCUUGUGGACCAGACCGAGCAGAAUGAAGGAAGGCCUGGGAAAGGCCGAGCAGACUGGUGGUCUGGAGCGGAGGGUCGGGCAUG .((((((..(((.((..((((((((..(((..((......(((((.....)))))...))..))).))))))))..)).))).))))))... ( -44.40) >consensus AGCCCGGAGCUUGUGGACCAGACCGAGCAGAAUGAAGGAAGGCCUGGGAAAGGCCGAGCAGACUGGUGGUCUGGAGCGGAGGGUCGGGCAUG .((((((..(((.((..((((((((..(((..((......(((((.....)))))...))..))).))))))))..)).))).))))))... (-40.90 = -41.57 + 0.67)


| Location | 881,576 – 881,668 |
|---|---|
| Length | 92 |
| Sequences | 3 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 96.01 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -29.87 |
| Energy contribution | -30.20 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 881576 92 - 27905053 CAUGCCCGACCCUCCGCUCCAGACCACCAGUCUGCCCGGCCGUUCCCAGGCCUUCCUUCAUUCUGCUCGGUCUGGUACACAAGCUCCGGGCU ...(((((...((.....(((((((..(((..((...((((.......))))......))..)))...)))))))......))...))))). ( -31.60) >DroSec_CAF1 49617 92 - 1 CAUGCCCGACCCUCCGCUCCAGACCACGAGUCUGCACGGCCUUUCCCAGGCCUUCCUUCAAUCUGCUCGGUCUGGUCCACAAGCUCCGGGCU ...(((((...((.....(((((((..((((.((...(((((.....)))))......))....)))))))))))......))...))))). ( -30.30) >DroSim_CAF1 48820 92 - 1 CAUGCCCGACCCUCCGCUCCAGACCACCAGUCUGCUCGGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGGUCUGGUCCACAAGCUCCGGGCU ...(((((...((.....(((((((..(((..((...(((((.....)))))......))..)))...)))))))......))...))))). ( -31.90) >consensus CAUGCCCGACCCUCCGCUCCAGACCACCAGUCUGCACGGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGGUCUGGUCCACAAGCUCCGGGCU ...(((((...((.....(((((((..(((..((...((((.......))))......))..)))...)))))))......))...))))). (-29.87 = -30.20 + 0.33)


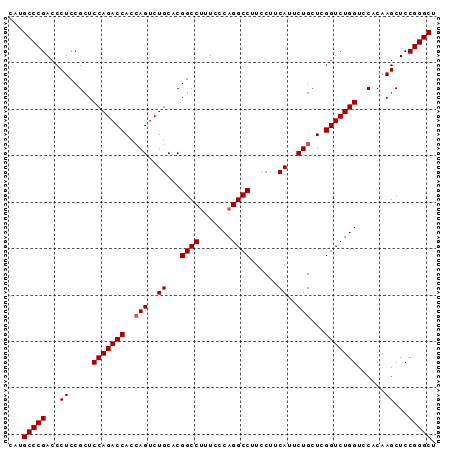
| Location | 881,598 – 881,708 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -28.39 |
| Energy contribution | -28.45 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.960322 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 881598 110 + 27905053 -------CCGAGCAGAAUGAAGGAAGGCCUGGGAACGGCCG--GGCAGACUGGUGGUCUGGAGCGGAGGG-UCGGGCAUGCCAAGUGGAAAUAAAGUAAUUUAAAUAUGAUGGCAGCACA -------((((.(............((((.(....))))).--..(((((.....)))))........).-))))((.(((((.(((....((((....))))..)))..)))))))... ( -33.80) >DroSec_CAF1 49639 110 + 1 -------CCGAGCAGAUUGAAGGAAGGCCUGGGAAAGGCCG--UGCAGACUCGUGGUCUGGAGCGGAGGG-UCGGGCAUGCCAAGUGGAAAUAAAGUAAUUUAAAUAUGAUGGCAGCACA -------((((.(............(((((.....)))))(--(.(((((.....)))))..))....).-))))((.(((((.(((....((((....))))..)))..)))))))... ( -36.20) >DroSim_CAF1 48842 110 + 1 -------CCGAGCAGAAUGAAGGAAGGCCUGGGAAAGGCCG--AGCAGACUGGUGGUCUGGAGCGGAGGG-UCGGGCAUGCCAAGUGGAAAUAAAGUAAUUUAAAUAUGAUGGCAGCACA -------((((.(............(((((.....))))).--..(((((.....)))))........).-))))((.(((((.(((....((((....))))..)))..)))))))... ( -34.80) >DroEre_CAF1 48618 116 + 1 ACGAAGACCGAACAGAA----CGAAGGCCUGGGAAUGGCCACCAGUAGACUGGUGGUCUGGAGCGGAGGGUUCGGGCACGCCAAGUGGAAAUAAAGUAAUUUAAAUAUGAUGGCAGCACA .......((((((....----((....((.......(((((((((....))))))))).))..))....))))))((..((((.(((....((((....))))..)))..)))).))... ( -37.70) >consensus _______CCGAGCAGAAUGAAGGAAGGCCUGGGAAAGGCCG__AGCAGACUGGUGGUCUGGAGCGGAGGG_UCGGGCAUGCCAAGUGGAAAUAAAGUAAUUUAAAUAUGAUGGCAGCACA .......((((.(............((((.......)))).....(((((.....))))).........).))))((.(((((.(((.((((......))))...)))..)))))))... (-28.39 = -28.45 + 0.06)

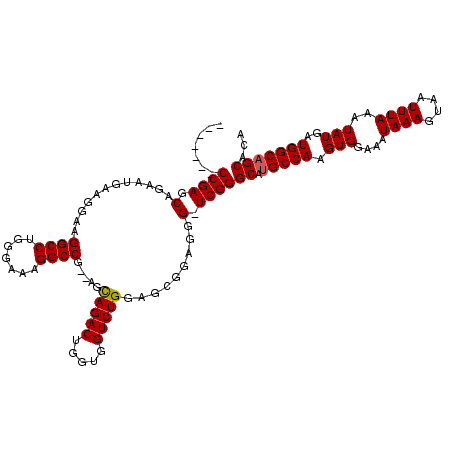
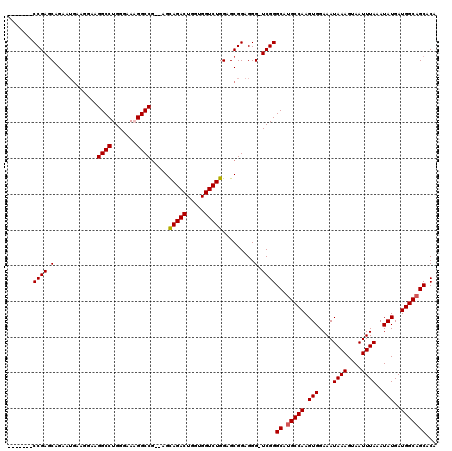
| Location | 881,598 – 881,708 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -26.08 |
| Consensus MFE | -22.67 |
| Energy contribution | -22.73 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 881598 110 - 27905053 UGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACCAGUCUGCC--CGGCCGUUCCCAGGCCUUCCUUCAUUCUGCUCGG------- ...(((((((.......((((....))))........))))).))((((-..........(((((.....)))))..--.((((.......))))..............))))------- ( -24.36) >DroSec_CAF1 49639 110 - 1 UGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACGAGUCUGCA--CGGCCUUUCCCAGGCCUUCCUUCAAUCUGCUCGG------- ...(((((((.......((((....))))........))))).))((((-..........(((((.....)))))..--.(((((.....)))))..............))))------- ( -24.66) >DroSim_CAF1 48842 110 - 1 UGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACCAGUCUGCU--CGGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGG------- ...(((((((.......((((....))))........))))).))((((-..........(((((.....)))))..--.(((((.....)))))..............))))------- ( -24.66) >DroEre_CAF1 48618 116 - 1 UGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCGUGCCCGAACCCUCCGCUCCAGACCACCAGUCUACUGGUGGCCAUUCCCAGGCCUUCG----UUCUGUUCGGUCUUCGU .(..(.((((.......((((....))))........)))))..).((((....(((...((((.((((((....)))))((((.......))))...)----.))))..)))..)))). ( -30.66) >consensus UGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA_CCCUCCGCUCCAGACCACCAGUCUGCU__CGGCCUUUCCCAGGCCUUCCUUCAUUCUGCUCGG_______ ...(((((((.......((((....))))........))))).))((((...........(((((.....))))).....((((.......))))..............))))....... (-22.67 = -22.73 + 0.06)

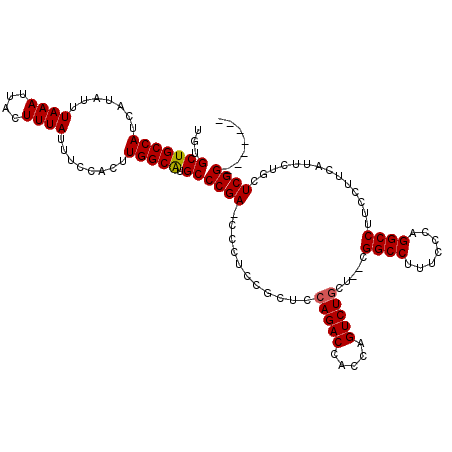
| Location | 881,631 – 881,748 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -27.84 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.46 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
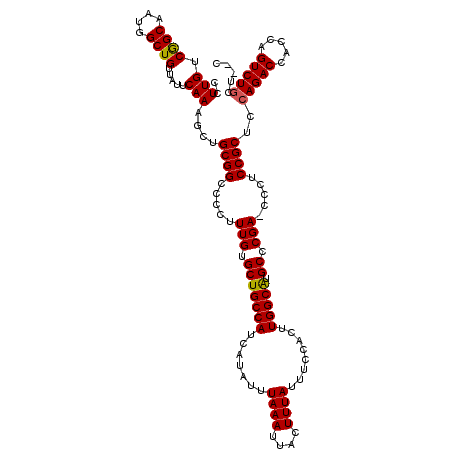
>3R_DroMel_CAF1 881631 117 - 27905053 CCUUGUCGGCAAUGGCUGUUAUUCAAAGCUGCGGCCCCUUUGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACCAGUCUGCC--C ....((((((((.((((((...........))))))...))).(((((((.......((((....))))........))))).))))))-).........(((((.....)))))..--. ( -27.86) >DroSec_CAF1 49672 117 - 1 CCUUGUCGGCAAUGGCUGUUAUUCAAAGCUGCGGCCCCUUUGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACGAGUCUGCA--C ....((((((((.((((((...........))))))...))).(((((((.......((((....))))........))))).))))))-).........(((((.....)))))..--. ( -27.86) >DroSim_CAF1 48875 117 - 1 CCUUGUCGGCAAUGGCUGUUAUUCAAAGCUGCGGCCCCUUUGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA-CCCUCCGCUCCAGACCACCAGUCUGCU--C ....((((((((.((((((...........))))))...))).(((((((.......((((....))))........))))).))))))-).........(((((.....)))))..--. ( -27.86) >DroEre_CAF1 48654 120 - 1 CCUUGUCAGCAAUUGCUGUUAUUCAAAGCUGCGGCCCCUUUGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCGUGCCCGAACCCUCCGCUCCAGACCACCAGUCUACUGGU ....((((((....)))..........(..((((....((((.(((((((.......((((....))))........))))).)).))))....))))..).))).(((((....))))) ( -27.76) >consensus CCUUGUCGGCAAUGGCUGUUAUUCAAAGCUGCGGCCCCUUUGUGCUGCCAUCAUAUUUAAAUUACUUUAUUUCCACUUGGCAUGCCCGA_CCCUCCGCUCCAGACCACCAGUCUGCU__C ..(((.((((....)))).....)))....((((.....(((.(((((((.......((((....))))........))))).)).))).....))))..(((((.....)))))..... (-24.58 = -24.46 + -0.12)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:50 2006