
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,129,715 – 6,129,889 |
| Length | 174 |
| Max. P | 0.684768 |

| Location | 6,129,715 – 6,129,815 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 78.75 |
| Mean single sequence MFE | -43.38 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
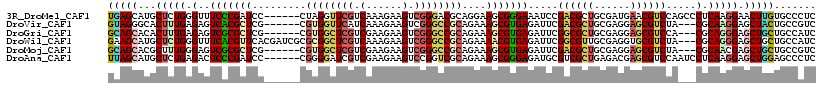
>3R_DroMel_CAF1 6129715 100 - 27905053 UGCGAUGAACGUCCAGCCCUCAAGGAACUUGUGCCCUCAGUGGCAG-----CAACUUGCGGCAAUUGCGGUGGAGGCGAGAGACAGCCACUCGGGGGACGUCCUA ((.(((....))))).......((((...(((.((((.((((((.(-----(...(..(.((....)).)..)..))(.....).)))))).)))).))))))). ( -33.20) >DroGri_CAF1 47861 100 - 1 UGCGAGGAGCGUCCA---CGCAGGGAGCUGCUGCCAUCGGUGACGG-UGG-CACCACCGGGUGCCUGUGGCGGCGGCGAUAGGUAGCCACUCGGUGGACGCCCCA .....((.(((((((---(((((....))))((((((((....)))-)))-)).....((.(((((((.((....)).))))))).)).....)))))))).)). ( -60.10) >DroSec_CAF1 41657 100 - 1 UGCGAGGAACGUCCAACCCUUAGGGAACUGGUGCCCUCAGUGGCAG-----CAACUUGCGGCAAUUGCGGUGGAGGCGAGAGACAGCCACUCGGAGGACGUCCUA ....((((.(((((.(((...((....)))))..((..((((((.(-----(...(..(.((....)).)..)..))(.....).)))))).)).))))))))). ( -35.30) >DroSim_CAF1 42701 100 - 1 UGCGAGGAACGUCCAACCCUCAGGGAACUUGUGCCCUCAGUGGCUG-----CAACUUGCGGCAAUUGCGGUGGAGGCGAGAGACAGCCACUCGGAGGACGUCCUA ....((((.(((((.......((((........))))((((.((((-----(.....))))).))))((((((..((....).)..)))).))..))))))))). ( -38.20) >DroEre_CAF1 42150 100 - 1 UGCGAGGAACGUCCAACCCUCAGGGAACUGGUGCCUUCAGUGGCAG-----CCACUUGUGGCAGUUGCGGUGGAGGCGAGAGGCAGCCACUCGGAGGACGUCCUA ....((((.(((((...((((((....)))).((((((((..((.(-----(((....)))).))..)..)))))))(((.((...)).))))).))))))))). ( -44.20) >DroWil_CAF1 44758 100 - 1 UGCGAGGUGCGUCUA---CGCAGGGAGCUGCUGCCAUCGG--GCAAACUGCCAAUGUUUGGCAAUUGCGGCGGCGGCGAGAGGCAACCACUUGGUGGACGCCCCA .....((.(((((((---(.((((..(((((((((.....--((((..(((((.....))))).)))))))))))))..(.(....)).)))))))))))).)). ( -49.30) >consensus UGCGAGGAACGUCCAACCCUCAGGGAACUGGUGCCCUCAGUGGCAG_____CAACUUGCGGCAAUUGCGGUGGAGGCGAGAGACAGCCACUCGGAGGACGUCCUA .....((.((((((...((((.....((((.((((...(((............)))...))))....)))).))))((((.(.....).))))..)))))))).. (-25.84 = -25.27 + -0.58)



| Location | 6,129,777 – 6,129,889 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 78.01 |
| Mean single sequence MFE | -45.97 |
| Consensus MFE | -29.68 |
| Energy contribution | -30.27 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6129777 112 - 27905053 UGAGCAUGCUCUGGGUUUCCCGAUCC------CUAGGUUCGUCAAAGAAGUCGGGACGCAGGAAGCGGGAAAUCCGACGCUGCGAUGAACGUCCAGCCCUCAAGGAACUUGUGCCCUC ...((((....(((((((((((...(------((.(..(((.(......).)))..)..)))...)))))))))))..((((.(((....)))))))((....)).....)))).... ( -36.20) >DroVir_CAF1 41458 109 - 1 GUAGGGCACUUUGAGAGUCACGCUCG------CGUGGUUCAUCAAAGAAGUCGGGCCGCAGAAAGCGUGAGAUUCGACGCUGCGAGGAGCGUCUA---CGCAAGGAGCUACUGCCGUC (((((((.(((((.((((((((((..------((((((((..(......)..))))))).)..)))))))..)))((((((......))))))..---..))))).))).)))).... ( -42.70) >DroGri_CAF1 47926 109 - 1 GCAGCACACUUUGAGAGUCGCGCUCG------CGUGGCUCGUCGAAGAAGUCGGGCCGCAGAAAGCGUGAGAUUCGGCGCUGCGAGGAGCGUCCA---CGCAGGGAGCUGCUGCCAUC (((((.......(((..(((((((..------(((((((((.(......).)))))))).)..)))))))..)))(((.(((((.((.....)).---)))))...)))))))).... ( -51.90) >DroWil_CAF1 44823 115 - 1 GAAGCAUGCUCUGGGUUUCACGUUCACGAUCGCGCGGCUCGUCAAAGAAGUCGGGCCGCAGAAAACGUGAGAUUCGGCGUUGCGAGGUGCGUCUA---CGCAGGGAGCUGCUGCCAUC ..((((.(((((((((((((((((.....((..((((((((.(......).)))))))).)).)))))))))))).((((.(((.....)))..)---)))..)))))))))...... ( -50.80) >DroMoj_CAF1 53803 109 - 1 GCAGCACGCUUUGGGAGUCGCGCUCG------CGUGGCUCGUCGAAGAAGUCGGGCCGCAGAAAGCGUGAGAUUCGACGCUGCGAGGAGCGUCUA---CGCAACGAGCUGCUGCCGUC ((((((.((((((.(((.(((.((((------(((((((((.(......).))))))))....(((((........))))))))))..))).)).---).))..)))))))))).... ( -54.60) >DroAna_CAF1 36933 112 - 1 UUAGCAUGCUCUGAGACUCCCGAUCC------CGGGGAUCGUCGAAGAAGUCCGGUCGCAGAAAGCGGGAGAUGCGUCGCUGAGACGAGCGUCCAAUCCUCAAGGAGCUGGAGCCCUC ...((.(((((.....(((((((..(------((((..((......))..)))))))((.....)))))))....(((.....))))))))((((.(((....)))..)))))).... ( -39.60) >consensus GCAGCACGCUCUGAGAGUCACGCUCG______CGUGGCUCGUCAAAGAAGUCGGGCCGCAGAAAGCGUGAGAUUCGACGCUGCGAGGAGCGUCCA___CGCAAGGAGCUGCUGCCAUC (((((...(((((.(..(((((((.........((((((((.(......).))))))))....))))))).....((((((......)))))).....).))))).)))))....... (-29.68 = -30.27 + 0.59)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:30 2006