
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,102,970 – 6,103,117 |
| Length | 147 |
| Max. P | 0.999888 |

| Location | 6,102,970 – 6,103,083 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.49 |
| Mean single sequence MFE | -29.62 |
| Consensus MFE | -19.14 |
| Energy contribution | -20.27 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990041 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6102970 113 + 27905053 UAACGGAAUCAUUU---UUCACAGUCUCCAAU-UUAAAUUAACUUCCGUGGGGUAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACC ..((((((......---...............-..........))))))(((((.(((((.((((((((((.......))))))))))..)))))..)))))............... ( -35.18) >DroSec_CAF1 15119 113 + 1 UAACGGAAACAUUU---UCUACAGUCUCCAAU-UUAAAUUAACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACC ..((((((......---...............-..........))))))((((..(((((.((((((((((.......))))))))))..)))))...))))............... ( -34.08) >DroSim_CAF1 15663 114 + 1 UAACGGAAACCUUU---UUCACAGUCUCCAGUUUUAAAUUAACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCNUUCUAACCACACACC ..((((((....((---(..((........))...))).....))))))(((...(((((.((((((((((.......))))))))))..)))))...)))................ ( -31.50) >DroEre_CAF1 15350 111 + 1 AAACGGAACUAUUC---UUC--AGUCUCCGAU-UUAAAUUAACUUCCGUGGGGAAAACCCUCUUAGCUGCAUUAAGGUUGCAGCUGAGCUGCGUUAAACCCCUUCUAACCACACACC ..((((((......---...--((((...)))-).........))))))((((..((((..((((((((((.......))))))))))..).)))...))))............... ( -30.17) >DroYak_CAF1 15506 113 + 1 CAACGGAACUAAUC---UUCACAGUCUCCCAU-UUAAAUUAACUUCCGUGGGGAAAACCCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACC ..((((((.((((.---...............-....))))..))))))((((..((((..((((((((((.......))))))))))..).)))...))))............... ( -27.85) >DroAna_CAF1 12175 97 + 1 UAAUGCAGCCAGUUCCCAUCUCAGUCCUACAG-GAACUUCAACUUGCAUCCUGAAAC----CUC-----------AACUGCGGCGGGGCACC----AACCUCCUCCAACCACAAGCC ....((.(((((((((...............)-))))).......(((...(((...----.))-----------)..))))))((((....----..))))............)). ( -18.96) >consensus UAACGGAACCAUUU___UUCACAGUCUCCAAU_UUAAAUUAACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACC ..((((((...................................))))))((((..(((...((((((((((.......))))))))))....)))...))))............... (-19.14 = -20.27 + 1.13)

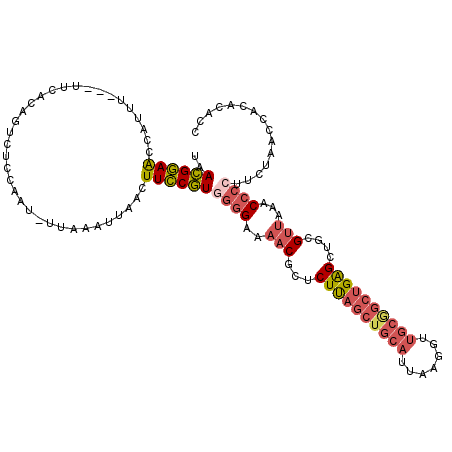
| Location | 6,103,006 – 6,103,117 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -26.62 |
| Energy contribution | -26.94 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.92 |
| SVM decision value | 4.39 |
| SVM RNA-class probability | 0.999888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6103006 111 + 27905053 AACUUCCGUGGGGUAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACAUAUCCAUUAUGU .........(((((.(((((.((((((((((.......))))))))))..)))))..)))))..................................(((((.....))))) ( -32.80) >DroSec_CAF1 15155 111 + 1 AACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACAUAUACAUUAUGC .........((((..(((((.((((((((((.......))))))))))..)))))...))))............................(((((...........))))) ( -32.00) >DroSim_CAF1 15700 111 + 1 AACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCNUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACAUAUACAUUAUGU .......(((((((((((((.((((((((((.......))))))))))..))))).......))))..))))........................(((((.....))))) ( -29.51) >DroEre_CAF1 15384 111 + 1 AACUUCCGUGGGGAAAACCCUCUUAGCUGCAUUAAGGUUGCAGCUGAGCUGCGUUAAACCCCUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACAUAUACAUUAUGU .........((((..((((..((((((((((.......))))))))))..).)))...))))..................................(((((.....))))) ( -26.20) >DroYak_CAF1 15542 111 + 1 AACUUCCGUGGGGAAAACCCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACUUAUACAUUGUAU .........((((..((((..((((((((((.......))))))))))..).)))...))))...........................((((.............)))). ( -23.82) >consensus AACUUCCGUGGGGAAAACGCUCUUAGCUGCAUUAAGGUUGCGGCUGAGCUGCGUUAAACCCCUUCUAACCACACACCAUCACCAUUAAUUGCAUAUACAUAUACAUUAUGU .........((((..(((((.((((((((((.......))))))))))..)))))...))))............................(((((...........))))) (-26.62 = -26.94 + 0.32)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:28:16 2006