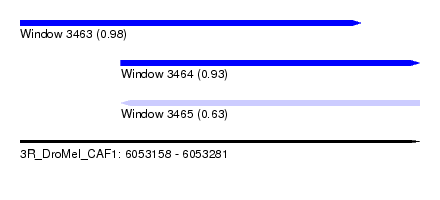
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,053,158 – 6,053,281 |
| Length | 123 |
| Max. P | 0.984249 |

| Location | 6,053,158 – 6,053,263 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.50 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -20.61 |
| Energy contribution | -21.20 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6053158 105 + 27905053 ACACACACGUGAGGUGGU-------ACCU-CUU-UUGGCCAUGGCUGAUAUGGCGAG------CUAUUGUUGGUGCCAUUGUUGCUGCAUUGUACGAGUGUGAUUCAUGUCAUGCUCUCA (((..((.(..(((((((-------(((.-...-...((((((.....)))))).((------(....)))))))))))).)..)))...)))..(((((((((....)))))))))... ( -34.20) >DroSec_CAF1 59060 92 + 1 -CACACACGUGAGAUGGU-------UCCU-CUU-UUGGCCAUGGCUGAUAUGGCCAG------CUAUU------------GUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA -.......(((.((((..-------....-...-..(((((((.....)))))))((------(....------------...))).)))).)))(((((((((....)))))))))... ( -31.40) >DroSim_CAF1 60386 93 + 1 -CACACACGUGAGGUAGU-------GCCU-CUUUUUGGCCAUGGCUGAUAUGGCCAG------CUAUU------------GUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA -.......(((..(((((-------..(.-.(..(((((((((.....)))))))))------..)..------------)..)))))....)))(((((((((....)))))))))... ( -34.30) >DroEre_CAF1 59475 101 + 1 ----ACGCCUCAUGUGAU-------GCCC-CUU-UUGGCCAUGGCUGAUAUGGCCAG------CCAUUGUUGCUGCCAUUGUUGCUGCAGUGUACGAGUGUGAUUCAUGUCACGCUCUCA ----..((.((....)).-------))..-...-(((((((((.....)))))))))------.((((((.((.((....)).)).))))))...(((((((((....)))))))))... ( -36.10) >DroYak_CAF1 61648 105 + 1 ACACACACAUGAGGUGAU-------GCCU-CUU-UUGGCCAUGACUGAUAUGGCCAG------CUAUUGUUGCUGCCAUUGUUGCUGCAGUGCACGAGUGUGAUUCAUGUCACGCUCUCA ..........((((....-------.)))-)..-.((((((((.....))))))))(------(.(((((.((.((....)).)).)))))))..(((((((((....)))))))))... ( -38.90) >DroAna_CAF1 53014 109 + 1 A--CCCACGGCAACUGCCCGCCCACCCAUCGUU-AUGGCCGUGUUUUGGAUGUGCAGAGGUGCCACUUUUUGCUGCCAUUGUUG--------UUUGAGUAUGAUUCAUGUCACGCUCGCA .--...(((((((((((.((.(((..(((.((.-...)).)))...))).)).)))).(((((........)).))).))))))--------)..(((..((((....))))..)))... ( -29.00) >consensus _CACACACGUGAGGUGGU_______GCCU_CUU_UUGGCCAUGGCUGAUAUGGCCAG______CUAUUGUUGCUGCCAUUGUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA ...................................((((((((.....)))))))).......................................(((((((((....)))))))))... (-20.61 = -21.20 + 0.59)



| Location | 6,053,189 – 6,053,281 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6053189 92 + 27905053 AUGGCUGAUAUGGCGAG------CUAUUGUUGGUGCCAUUGUUGCUGCAUUGUACGAGUGUGAUUCAUGUCAUGCUCUCA--CACUCGACCAGCGAAAAG .(((((.........))------)))((((((((..((.(((....))).))..(((((((((..(((...)))...)))--)))))))))))))).... ( -28.40) >DroSec_CAF1 59090 80 + 1 AUGGCUGAUAUGGCCAG------CUAUU------------GUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA--CACUCGACCAGCGAAAAG (((((((.......)))------)))).------------.((((((...(((..(((((((((....)))))))))..)--))......)))))).... ( -27.10) >DroSim_CAF1 60417 80 + 1 AUGGCUGAUAUGGCCAG------CUAUU------------GUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA--CACUCGACCAGCGAAAAG (((((((.......)))------)))).------------.((((((...(((..(((((((((....)))))))))..)--))......)))))).... ( -27.10) >DroEre_CAF1 59502 92 + 1 AUGGCUGAUAUGGCCAG------CCAUUGUUGCUGCCAUUGUUGCUGCAGUGUACGAGUGUGAUUCAUGUCACGCUCUCA--CACUCGACCAGCGAAAAG (((((((.......)))------))))..((((((.....((((....(((((..(((((((((....)))))))))..)--)))))))))))))).... ( -35.10) >DroYak_CAF1 61679 92 + 1 AUGACUGAUAUGGCCAG------CUAUUGUUGCUGCCAUUGUUGCUGCAGUGCACGAGUGUGAUUCAUGUCACGCUCUCA--CACUCGACCAGCGAAAAG ..(((....(((((.((------(.......)))))))).)))((((.((((...(((((((((....)))))))))...--))))....))))...... ( -32.50) >DroAna_CAF1 53051 92 + 1 GUGUUUUGGAUGUGCAGAGGUGCCACUUUUUGCUGCCAUUGUUG--------UUUGAGUAUGAUUCAUGUCACGCUCGCAAGCACUCGCCCUGCAAAAAG .(((...((..((((...(((((........)).)))....(((--------(..(((..((((....))))..)))))))))))...))..)))..... ( -24.10) >consensus AUGGCUGAUAUGGCCAG______CUAUUGUUGCUGCCAUUGUUGCUGCAUUGUACGAGUGUGAUUCAUGUCACGCUCUCA__CACUCGACCAGCGAAAAG .(((((.....))))).........................((((((........(((((((((....)))))))))((........)).)))))).... (-19.55 = -19.97 + 0.42)



| Location | 6,053,189 – 6,053,281 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.98 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -12.16 |
| Energy contribution | -12.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.625152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6053189 92 - 27905053 CUUUUCGCUGGUCGAGUG--UGAGAGCAUGACAUGAAUCACACUCGUACAAUGCAGCAACAAUGGCACCAACAAUAG------CUCGCCAUAUCAGCCAU ......((((((((((((--(((...(((...)))..))))))))).))............(((((...........------...)))))..))))... ( -27.14) >DroSec_CAF1 59090 80 - 1 CUUUUCGCUGGUCGAGUG--UGAGAGCGUGACAUGAAUCACACUCGUACAAUGCAGCAAC------------AAUAG------CUGGCCAUAUCAGCCAU ......((((((((((((--(((...(((...)))..))))))))).))....((((...------------....)------))).......))))... ( -26.40) >DroSim_CAF1 60417 80 - 1 CUUUUCGCUGGUCGAGUG--UGAGAGCGUGACAUGAAUCACACUCGUACAAUGCAGCAAC------------AAUAG------CUGGCCAUAUCAGCCAU ......((((((((((((--(((...(((...)))..))))))))).))....((((...------------....)------))).......))))... ( -26.40) >DroEre_CAF1 59502 92 - 1 CUUUUCGCUGGUCGAGUG--UGAGAGCGUGACAUGAAUCACACUCGUACACUGCAGCAACAAUGGCAGCAACAAUGG------CUGGCCAUAUCAGCCAU ......((((((..((((--((.(((.((((......)))).))).)))))).........((((((((.......)------)).)))))))))))... ( -31.90) >DroYak_CAF1 61679 92 - 1 CUUUUCGCUGGUCGAGUG--UGAGAGCGUGACAUGAAUCACACUCGUGCACUGCAGCAACAAUGGCAGCAACAAUAG------CUGGCCAUAUCAGUCAU ......((((((((((((--(((...(((...)))..)))))))))(((......)))...((((((((.......)------)).)))))))))))... ( -30.60) >DroAna_CAF1 53051 92 - 1 CUUUUUGCAGGGCGAGUGCUUGCGAGCGUGACAUGAAUCAUACUCAAA--------CAACAAUGGCAGCAAAAAGUGGCACCUCUGCACAUCCAAAACAC .....(((((((...(((((((((((..(((......)))..)))...--------((....))...)))......))))))))))))............ ( -22.30) >consensus CUUUUCGCUGGUCGAGUG__UGAGAGCGUGACAUGAAUCACACUCGUACAAUGCAGCAACAAUGGCAGCAACAAUAG______CUGGCCAUAUCAGCCAU ......((((.(((......)))(((.((((......)))).)))........))))...........................((((.......)))). (-12.16 = -12.83 + 0.67)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:54 2006