
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,048,533 – 6,048,794 |
| Length | 261 |
| Max. P | 0.645231 |

| Location | 6,048,533 – 6,048,632 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.59 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.98 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.595347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6048533 99 - 27905053 CUCCUCGCUGGAGUUCGAUGCAGCCCUAUUAUCCGUAUCAAGAUCGUAAGAUUGUAGCUGAUAGCUGGCUGCAUUGGCGAGAUGUAGGGAACAGAUCAA ((((.....)))).(((((((((((...(((((.((.....((((....))))...)).)))))..))))))))))).((..(((.....)))..)).. ( -34.30) >DroSec_CAF1 54461 99 - 1 CUCUCCGCUGGAGUUCGAUGCAGCCCCAUUAUCCGCAUCAAGAUCGUAAGAUUGUAGCUGAUAGCUGGCUGCAUGGGCAAGAUCUAGGGAACAGAUCAA ..((((...)))).((.((((((((...(((((.((.....((((....))))...)).)))))..)))))))).))...(((((.(....)))))).. ( -35.40) >DroSim_CAF1 55712 99 - 1 CUGCUCGCUGGAGUUCGAUGCAGCCCCAUUAUCCGCAUUAAGAUCGUAAGAUUGUAGCUGAAUGCUGGCUGCAUACGCAAAAUCUAGGGAACAGAUCAA (((.((.(((((...((((((((((.((((....((.....((((....))))...))..))))..)))))))).)).....))))).)).)))..... ( -34.70) >DroEre_CAF1 55010 99 - 1 CUCCUCGCUAGAGUUCGAUACAGCCGCGUUAUCCGUAUCCAGAUCGUAGGAUUGCAGCUGAUAACUGGCUGCAUUUGCGAGAUCUAGGUAACAGAUAAA .......(((((.((((...((((((.((((((.((((((........))))....)).))))))))))))......)))).)))))............ ( -29.00) >DroYak_CAF1 56941 99 - 1 CUCCUCGCUGGAGUUCGAUACAGCUCCAUUAUCCGUAUCCAGGUCGUAGGAUUGCAGCUGAUAGCUGGCUGCAUUUGCGAGAUCUAGGUACCAGAUCAA ....(((((((((((......)))))))........((((........))))(((((((.......)))))))...))))(((((.(....)))))).. ( -31.00) >DroPer_CAF1 53471 99 - 1 GCCUUCGGUGCUGUUUCCUGACACUUCAUUAUCCAUAUCCAAGUCGUAUAUCUGCAGCUGUCUGCUGGCUGCUUCGCCCAGACCGAGGAAGCAGACUAG ......(((.(((((((((......................(((.(((....))).)))(((((..(((......))))))))..)))))))))))).. ( -27.10) >consensus CUCCUCGCUGGAGUUCGAUGCAGCCCCAUUAUCCGUAUCAAGAUCGUAAGAUUGCAGCUGAUAGCUGGCUGCAUUGGCGAGAUCUAGGGAACAGAUCAA ............(((((((((.............))))).(((((.......(((((((.......))))))).......)))))...))))....... (-16.58 = -16.98 + 0.39)

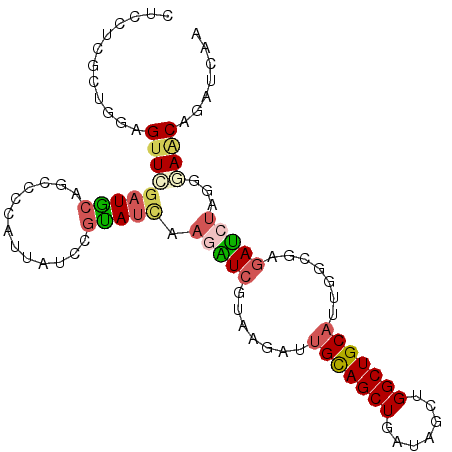

| Location | 6,048,598 – 6,048,695 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -18.39 |
| Energy contribution | -20.25 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645231 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6048598 97 - 27905053 GCUGUUGGCAUUCCGCAGCCAAGAAAGAGGUCUUUCCAGGGACGUGACUCCGC------CAUUGAACACCUCCUCGCUGGAGUUCGAUGCAGCCCUAUUAUCC (((((.(((....(((..((..(((((....)))))...))..))).....))------)(((((((.((........)).)))))))))))).......... ( -29.00) >DroPse_CAF1 53340 100 - 1 GCUCUGGACCUGCCGCAGCCAGCUCAGAGGUCUCUCCAUC---GUCAGCCCAGAACUGGGAUUGUACUGGCCUUCGGUGCUGUUUCCUGACACUUCAUUAUCC .(((((..(..((....))..)..)))))...........---(((((((((....))))...((((((.....))))))......)))))............ ( -27.80) >DroSec_CAF1 54526 97 - 1 GCUGUUGGCAUUUCGUAGCCAAGAAAGAGGUCUUUCCAGAGAUGUUACUUCGC------CAUUGAACUCCUCUCCGCUGGAGUUCGAUGCAGCCCCAUUAUCC (((((((((........)))))(((.((.(((((....))))).))..)))..------(((((((((((........))))))))))))))).......... ( -32.00) >DroSim_CAF1 55777 97 - 1 GCUGUUGGCAUUCCGUAGCCAAGAAAGAGGUCUUUCCAGAGAUGUCACUUCGC------CAUUGAACUCCUGCUCGCUGGAGUUCGAUGCAGCCCCAUUAUCC (((((((((........)))))(((.((.(((((....))))).))..)))..------(((((((((((........))))))))))))))).......... ( -33.80) >DroEre_CAF1 55075 103 - 1 GCUGUUGGCGUUCCGUAGCCAGGAAAGAGGUCUUUCCAGGGCGGUGACUCCUGCACUUGCAUUGAACUCCUCCUCGCUAGAGUUCGAUACAGCCGCGUUAUCC (((((.((.(((((((..((.((((((....)))))).)))))).))).)).((....))(((((((((..........)))))))))))))).......... ( -34.40) >DroYak_CAF1 57006 97 - 1 GCUGUUGGCGUUCCGUAGCCAAGAAAGAGGUCUUUCCAGGGAGGUAACUCCUG------CAUUGAACUCCUCCUCGCUGGAGUUCGAUACAGCUCCAUUAUCC (((((((((........)))).(((((....)))))..(((((....))))).------.((((((((((........))))))))))))))).......... ( -36.30) >consensus GCUGUUGGCAUUCCGUAGCCAAGAAAGAGGUCUUUCCAGGGAGGUCACUCCGC______CAUUGAACUCCUCCUCGCUGGAGUUCGAUGCAGCCCCAUUAUCC (((((((((........)))).(((((....)))))........................((((((((((........))))))))))))))).......... (-18.39 = -20.25 + 1.86)

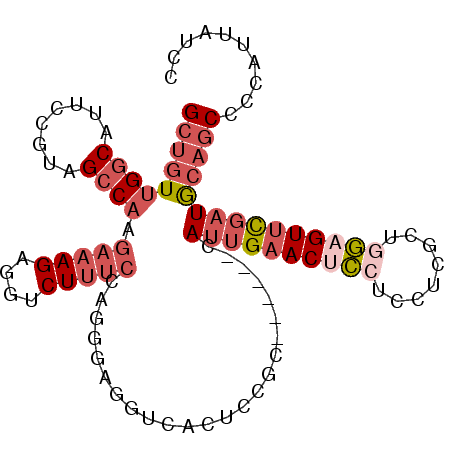

| Location | 6,048,695 – 6,048,794 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -28.90 |
| Energy contribution | -28.10 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6048695 99 + 27905053 GUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGUUACAUCGAUCAGCCGGAAACAGUCAAGUGGGUUAA---------GGAUACA--UUACC (((((((((((..((..(((((((.(((....))).))))))).))..)))))..(((.((..((.(....).))...)).)))...---------)))))).--..... ( -36.00) >DroSec_CAF1 54623 108 + 1 AUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGCUACAUCGAUCAGCCGGAAACAGUCAAGUGGGUUAAGGAUAUCAAGGAUACU--UUCCC (((((((((((..((..(((((((.(((....))).))))))).))..)))))..(((.((..((.(....).))...)).)))...))))))...(((....--.))). ( -37.50) >DroSim_CAF1 55874 108 + 1 AUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGCUACAUCGAUCAGCCGGAAACAGUCAAGUGGGUUAAAGAUAUCAAGGAUACU--UUCCC .((((((((((..((..(((((((.(((....))).))))))).))..)))))..(((....(((((....)...(....)))))...))).....)))))..--..... ( -36.10) >DroEre_CAF1 55178 103 + 1 AUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGCUGCAUCGCUCAGCCGGAAACAGUCAGGUGGGCUAAGGACAUCAAGGCUGGA------- .....((((((..((..(((((((.(((....))).))))))).))..))))))......(((((((....).(((.((....))...))).....)))))).------- ( -44.90) >DroYak_CAF1 57103 108 + 1 AUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGCUGCAUCGCUCAGCCGGGAACAGUCAAGUGGGUUGAGAAUAUCAAGGCUACA--UUCCC ((((.((((((..((..(((((((.(((....))).))))))).))..)))))).....(((((((....((......)).)))))))............)))--).... ( -40.00) >DroMoj_CAF1 69440 101 + 1 AUGUUCGCCAGCCCGGCGGGGCAUGUGGUCGUCCAGGUGGCCAAGGAGCUGCUGCAUCGCUCCGCGGGCAAUAGUCAAGUGCGUGCG------CAGU-UUGCAUAUUA-- ..........(((((.((((((((((((....)).((..((......))..)))))).)))))))))))((((((.((.(((....)------)).)-).)).)))).-- ( -39.40) >consensus AUGUUCGGCAGUCCCGCUGGUCAUGUGGUCAUCCAGGUGGCCAAGGAGCUGCUACAUCGAUCAGCCGGAAACAGUCAAGUGGGUUAAGGAUAUCAAGGAUACA__UUCCC ((((..((((((.((..(((((((.(((....))).))))))).)).)))))))))).....((((....((......)).))))......................... (-28.90 = -28.10 + -0.80)


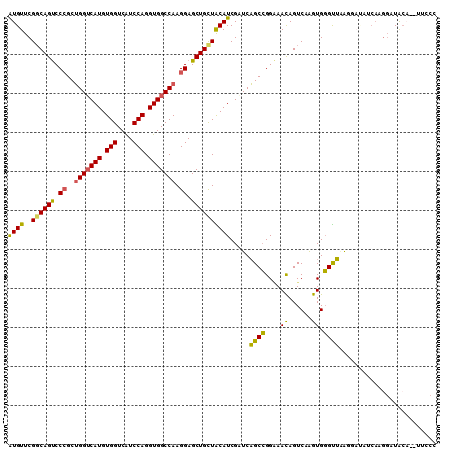
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:51 2006