
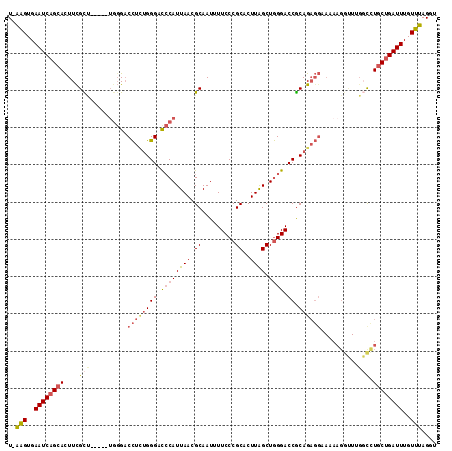
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 6,018,699 – 6,018,808 |
| Length | 109 |
| Max. P | 0.569666 |

| Location | 6,018,699 – 6,018,808 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -25.56 |
| Energy contribution | -26.32 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.525030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6018699 109 + 27905053 U-AAGUGAAUCAGCACUUUGCU-----UGCGACCUUUGGGACACAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGCAAAAGGCUUGGCCUGCUGAUUUGUUUAGGU (-(((..(((((((.(((((((-----((((.....((.....))....)))))...((((((.....)).))))..)))))).....((((...)))))))))))..))))... ( -38.60) >DroSec_CAF1 30493 109 + 1 U-AAGUGAAUCAGCACUUCGCU-----UGGGACCUUUGGGACCCAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGAAAAAGGUUUGGCUUGCUGAUUUGUUUAGGU (-(((..((((((((((..(((-----(....((((((((.((((((((.((.........))..)))).)))).)).))))))....))))..))..))))))))..))))... ( -35.90) >DroSim_CAF1 30627 109 + 1 U-AAGUGAAUCAGCACUUCGCU-----UGGGACCUCUGGGACCCAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGAAAAAGGUUUGGCCUGCUGAUUUGUUUAGGU (-(((..((((((((((..(((-----(....((((((((.((((((((.((.........))..)))).)))).)).))))))....))))..))..))))))))..))))... ( -38.50) >DroEre_CAF1 30722 109 + 1 U-AAGUGAAUCAGCACUUCGUU-----UGGGACCUCUGGGACCCAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGAAAAAGGUUGGCUUUGCUGAUUUGUUUAGGU (-(((..(((((((.(((((((-----((((.((....)).))))..)))((.....((((((.....)).))))..)).))))...((((....)))))))))))..))))... ( -36.00) >DroYak_CAF1 32451 109 + 1 U-AAGUGAAUCAGCACUUCGCC-----GUGGACCUCUGGGACCCAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGAAAAAGGUUGGGCCUGCUGAUUUGUUUAGGU (-(((..((((((((((..(((-----.....((((((((.((((((((.((.........))..)))).)))).)).)))))).....)))..))..))))))))..))))... ( -40.50) >DroAna_CAF1 27522 100 + 1 UUUUGUGAAUCAGAAUUUCGUAUGCCCUGGGUCUCGGGGGAUCCAUUAACGCAAUUUUCCCGCACUUGGCUAGGACCACAG---------------CCUGCCGAUUUGUAAAGGU (((((..((((((((((.(((......(((((((....)))))))...))).)))))).........(((.(((.......---------------))))))))))..))))).. ( -27.30) >consensus U_AAGUGAAUCAGCACUUCGCU_____UGGGACCUCUGGGACCCAUUAACGCAAUUUUCCCGCACUUAGCUGGGACCGCAGAGGAAAAAGGUUUGGCCUGCUGAUUUGUUUAGGU ..(((..((((((((....(((..........((((((((.((((((((.((.........))..)))).)))).)).))))))..........))).))))))))..))).... (-25.56 = -26.32 + 0.76)


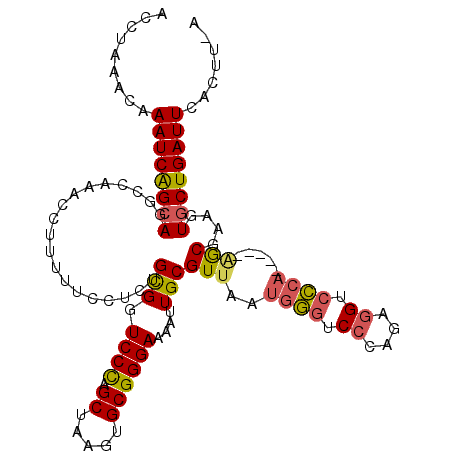
| Location | 6,018,699 – 6,018,808 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 84.14 |
| Mean single sequence MFE | -31.67 |
| Consensus MFE | -24.39 |
| Energy contribution | -24.37 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569666 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6018699 109 - 27905053 ACCUAAACAAAUCAGCAGGCCAAGCCUUUUGCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGUGUCCCAAAGGUCGCA-----AGCAAAGUGCUGAUUCACUU-A .........(((((((((((...)))((((((...((((..(...((.....))((((..((((...))))...))))...)..))))-----.)))))))))))))).....-. ( -33.40) >DroSec_CAF1 30493 109 - 1 ACCUAAACAAAUCAGCAAGCCAAACCUUUUUCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGGGUCCCAAAGGUCCCA-----AGCGAAGUGCUGAUUCACUU-A .........((((((((.((................(((.((((.((.....))))))....))).....((((.((....)).))))-----.))....)))))))).....-. ( -31.00) >DroSim_CAF1 30627 109 - 1 ACCUAAACAAAUCAGCAGGCCAAACCUUUUUCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGGGUCCCAGAGGUCCCA-----AGCGAAGUGCUGAUUCACUU-A .........((((((((((.....)).....((((((.((.((((..(((.(((((.....)))))))).)))).)))))))).....-----.......)))))))).....-. ( -34.70) >DroEre_CAF1 30722 109 - 1 ACCUAAACAAAUCAGCAAAGCCAACCUUUUUCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGGGUCCCAGAGGUCCCA-----AACGAAGUGCUGAUUCACUU-A .........((((((((..............((((((.((.((((..(((.(((((.....)))))))).)))).)))))))).....-----.......)))))))).....-. ( -34.47) >DroYak_CAF1 32451 109 - 1 ACCUAAACAAAUCAGCAGGCCCAACCUUUUUCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGGGUCCCAGAGGUCCAC-----GGCGAAGUGCUGAUUCACUU-A .........((((((((.(((..........((((((.((.((((..(((.(((((.....)))))))).)))).)))))))).....-----)))....)))))))).....-. ( -35.86) >DroAna_CAF1 27522 100 - 1 ACCUUUACAAAUCGGCAGG---------------CUGUGGUCCUAGCCAAGUGCGGGAAAAUUGCGUUAAUGGAUCCCCCGAGACCCAGGGCAUACGAAAUUCUGAUUCACAAAA .........((((((..((---------------(((......)))))..((((((((..((((...))))...))))((........))))))........))))))....... ( -20.60) >consensus ACCUAAACAAAUCAGCAGGCCAAACCUUUUUCCUCUGCGGUCCCAGCUAAGUGCGGGAAAAUUGCGUUAAUGGGUCCCAGAGGUCCCA_____AGCGAAGUGCUGAUUCACUU_A .........((((((((...................(((.((((.((.....))))))....)))(((..((((.((....)).)))).....)))....))))))))....... (-24.39 = -24.37 + -0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:37 2006