| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
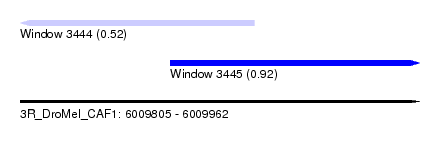
| Location | 6,009,805 – 6,009,962 |
| Length | 157 |
| Max. P | 0.920272 |

| Location | 6,009,805 – 6,009,897 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.62 |
| Mean single sequence MFE | -26.05 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.520484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6009805 92 - 27905053 ACCAAUACACUUGGCAAAUAA-CCGAGCCACUG---AGUUGGCGCAAUUUAACGCCGCGUCGAUCCAGCAGUGCGAAUUAUUUCGCUCAAUGCUGG------- .........(((((.......-)))))......---.((((((((...........))))))))((((((..(((((....)))))....))))))------- ( -28.70) >DroPse_CAF1 20604 99 - 1 ----AUAUAGUUUGUAUACAAUUCCAGCCACUCGGAGGCUGACGCAAUUUAACGCCGCGUUGCUUUUGCCGUGCAAAUUAUUUCGCCCAAUGCUAAUGCCUCU ----..(((((((((((.......(((((.......)))))..((((..(((((...)))))...)))).)))))))))))..........((....)).... ( -22.70) >DroSec_CAF1 21736 92 - 1 ACCAAUACACUUGGCAAAUAA-CCGAGCCACUG---AGUUGGCGCAAUUUAACGCCGCGUUGAUCCAGCAGUGCGAAUUAUUUUGCUCAAUGCUGA------- ..(((..(.(((((.......-)))))......---....((((........)))))..)))...(((((..(((((....)))))....))))).------- ( -24.50) >DroSim_CAF1 21748 92 - 1 ACCAAUACACUUGGCAAAUAA-CCGAGCCACUG---AGUUGGCGCAAUUUAACGCCGCGUUGAUCCAGCAGUGCGAAUUAUUUUGCUCAAUGCUGA------- ..(((..(.(((((.......-)))))......---....((((........)))))..)))...(((((..(((((....)))))....))))).------- ( -24.50) >DroEre_CAF1 22015 92 - 1 ACCAAUACACUUGGCAAAUAA-CCGAGCCACUG---AGUUGGCGCAAUUUAACGCAGCGCUGAUCCAGCAGUGCGAAUUAUUUUGCUCAAUGCUAG------- ...........(((((.....-....((((...---...))))((((..(((..(.((((((......)))))))..)))..))))....))))).------- ( -25.60) >DroYak_CAF1 23555 92 - 1 ACCAAUUCAGUUGGCAAAUAA-CUGAGCCACUG---AGUUGGCGCAAUUUAACGCGGCGCUGAUCCAGCAGUGCGAAUUAUUUUACUCAAUGCUGG------- .((((((((((.(((......-....)))))))---))))))(((........)))((((((......))))))......................------- ( -30.30) >consensus ACCAAUACACUUGGCAAAUAA_CCGAGCCACUG___AGUUGGCGCAAUUUAACGCCGCGUUGAUCCAGCAGUGCGAAUUAUUUUGCUCAAUGCUGA_______ ...........((((...........)))).......((((((((...........))))))))((((((..(((((....)))))....))))))....... (-18.27 = -18.63 + 0.36)



| Location | 6,009,864 – 6,009,962 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 69.60 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -12.05 |
| Energy contribution | -13.67 |
| Covariance contribution | 1.62 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 6009864 98 + 27905053 UCAGUGGCUCGGUUAUUUGCCAAGUGUAUUGGUAUUGCUCUGAUCCAACAGAGCUAGUGGCUCGGCA-----GUGAUGCCAGCAAUAUCAA---AAAACACUA---AUG---- ....((((..........))))(((((.(((((((((((...........((((.....))))((((-----....)))))))))))))))---...))))).---...---- ( -35.70) >DroSec_CAF1 21795 98 + 1 UCAGUGGCUCGGUUAUUUGCCAAGUGUAUUGGUAUUACUCCGGUCUCACAGAGCUUGUGGCUCGGCA-----GUGAUGCCAGCAAUAUCAA---AAAUCAGUA---AUC---- ...((((..(((.....((((((.....)))))).....)))..).))).((((.....))))((((-----....))))...........---.........---...---- ( -29.20) >DroSim_CAF1 21807 98 + 1 UCAGUGGCUCGGUUAUUUGCCAAGUGUAUUGGUAUUACUCCGGUCACACAGAGCUAGUGGCUCGGCA-----GUGAUGCCAGCAAUAUCAA---AAAACAGUA---AUC---- .((.(((((((((.....)))..(((((((((.......))))).)))).)))))).))(((.((((-----....)))))))........---.........---...---- ( -34.60) >DroEre_CAF1 22074 100 + 1 UCAGUGGCUCGGUUAUUUGCCAAGUGUAUUGGUAU--CUAUUGUCCCACAGAGCUGGUAGCUCGCCA-----GUGAUACCAGCGAUAUCAA---AAAAGAAUA---AAAUAUG ...((((..((((....((((((.....)))))).--..))))..))))...(((((((..(((...-----.))))))))))........---.........---....... ( -27.40) >DroYak_CAF1 23614 106 + 1 UCAGUGGCUCAGUUAUUUGCCAACUGAAUUGGUAUUGCUAUAAUCCCACAGAGCUGCUAGCUUGAUAGUGUAGUGAUGCCAGCAAUAUCAA---AAACAA-AA---ACCCUUU ((((((((..........))).))))).(((((((..(((((........((((.....)))).....)))))..))))))).........---......-..---....... ( -29.62) >DroAna_CAF1 19584 85 + 1 GCAGU-------------GCCAGCCGGAUUGGUU------UGAUCUCACAGCUUAAUAGACAUAAUA-----GUGAUUCCAUCAAUCUACAGAGAAAACACGAUAAAUU---- .((((-------------.((....))))))(((------(..((((...(((....)).)......-----((((((.....)))).)).))))))))..........---- ( -9.60) >consensus UCAGUGGCUCGGUUAUUUGCCAAGUGUAUUGGUAUU_CUCUGAUCCCACAGAGCUAGUGGCUCGGCA_____GUGAUGCCAGCAAUAUCAA___AAAACACUA___AUC____ ....((((..........))))......((((((((((............((((.....)))).........))))))))))............................... (-12.05 = -13.67 + 1.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:35 2006