
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,998,192 – 5,998,299 |
| Length | 107 |
| Max. P | 0.999959 |

| Location | 5,998,192 – 5,998,299 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 68.77 |
| Mean single sequence MFE | -22.61 |
| Consensus MFE | -13.59 |
| Energy contribution | -15.90 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5998192 107 + 27905053 GGAAUUUUAUUGUAUACGCAGAGAAAAAGAGGUUCCUGUACUC-----AACCAUAUUAUGAACUCUGAUUAAACUCUGAUUUAUCUCUGUGUACCUAUAAACGUCAGAUUUA .((..(((((....((((((((((...((((.(((..(((...-----.....)))...)))))))(((((.....)))))..))))))))))...)))))..))....... ( -20.30) >DroSec_CAF1 10158 107 + 1 GGGAUUUCAUAGGAUGCACAGAAAAAAAGAGGUUCCUGUACUC-----AACCAUAUUAUGAACUCUGAUUAAACUCUUAUUUAUCUCUGUGCACCUAUAAACGUCAGAUUUA ..(((((.(((((.(((((((((((.(((((((((..(((...-----.....)))...))))..........))))).)))...))))))))))))).)).)))....... ( -23.00) >DroSim_CAF1 8814 81 + 1 GCUAUUUCAUAGGGUACACAGAGAAAAAGAGGUUCCU-----------------------------GAUUAA--UCUGAUUUAUCUCUGUGCACCUAUAAACGUCAGAUUUA ........(((((...((((((((.(((.(((((...-----------------------------....))--)))..))).))))))))..))))).............. ( -20.30) >DroYak_CAF1 11579 96 + 1 GGUAAUUUAUAAGAUGCACAGAGAAAAAGUUGUUCUCAUACUAGAUAUAGCAAUUCUAUGA--------------CUGGUU--UCUCUGUGCAGCUAUAAACGUCGGAUUUA .....((((((...((((((((((((.(((..(.......(((....))).......)..)--------------))..))--))))))))))..))))))........... ( -26.84) >consensus GGUAUUUCAUAGGAUACACAGAGAAAAAGAGGUUCCUGUACUC_____AACCAUAUUAUGA_____GAUUAA__UCUGAUUUAUCUCUGUGCACCUAUAAACGUCAGAUUUA .....((((((((.((((((((((...((((..........................................))))......))))))))))))))))))........... (-13.59 = -15.90 + 2.31)



| Location | 5,998,192 – 5,998,299 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 68.77 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -16.93 |
| Energy contribution | -16.49 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.70 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5998192 107 - 27905053 UAAAUCUGACGUUUAUAGGUACACAGAGAUAAAUCAGAGUUUAAUCAGAGUUCAUAAUAUGGUU-----GAGUACAGGAACCUCUUUUUCUCUGCGUAUACAAUAAAAUUCC .......((..(((((..((((.((((((......((((((((((((............)))))-----))).........))))...)))))).))))...)))))..)). ( -18.90) >DroSec_CAF1 10158 107 - 1 UAAAUCUGACGUUUAUAGGUGCACAGAGAUAAAUAAGAGUUUAAUCAGAGUUCAUAAUAUGGUU-----GAGUACAGGAACCUCUUUUUUUCUGUGCAUCCUAUGAAAUCCC ...........((((((((((((((((((.....(((((((((((((............)))))-----))).........)))))..)))))))))).))))))))..... ( -29.90) >DroSim_CAF1 8814 81 - 1 UAAAUCUGACGUUUAUAGGUGCACAGAGAUAAAUCAGA--UUAAUC-----------------------------AGGAACCUCUUUUUCUCUGUGUACCCUAUGAAAUAGC .....(((...((((((((((((((((((.(((...((--....))-----------------------------((....)).))).)))))))))).)))))))).))). ( -23.10) >DroYak_CAF1 11579 96 - 1 UAAAUCCGACGUUUAUAGCUGCACAGAGA--AACCAG--------------UCAUAGAAUUGCUAUAUCUAGUAUGAGAACAACUUUUUCUCUGUGCAUCUUAUAAAUUACC ..........((((((((.((((((((((--((..((--------------(........(((((....)))))........))).))))))))))))..)))))))).... ( -25.19) >consensus UAAAUCUGACGUUUAUAGGUGCACAGAGAUAAAUCAGA__UUAAUC_____UCAUAAUAUGGUU_____GAGUACAGGAACCUCUUUUUCUCUGUGCAUCCUAUAAAAUACC ...........((((((((((((((((((...........................................................))))))))))).)))))))..... (-16.93 = -16.49 + -0.44)

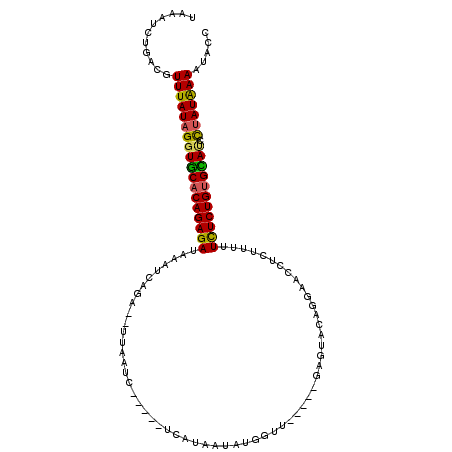
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:30 2006