| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
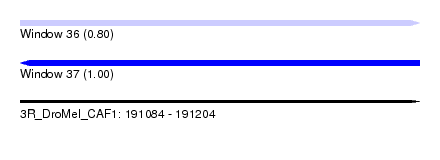
| Location | 191,084 – 191,204 |
| Length | 120 |
| Max. P | 0.997618 |

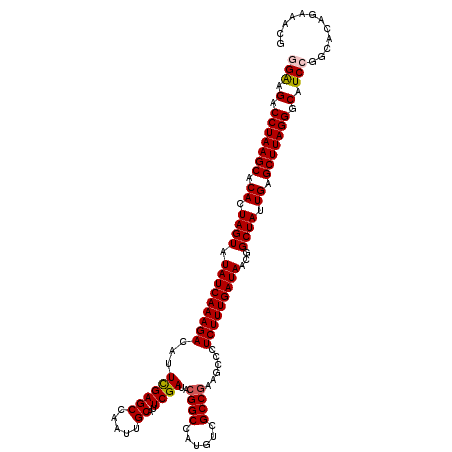
| Location | 191,084 – 191,204 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -39.29 |
| Consensus MFE | -34.22 |
| Energy contribution | -34.80 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.797587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 191084 120 + 27905053 CGUGUCUGUCCCGGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUUUGGCGACAUGGCCGUGUCGAACGCUAUAGGCUCGAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUUCC .(((((((...)))))))(((((((.((.(((....(((((((((.((((.((....)).))))...((((..((.....))))))...)))))))))..))).)).)))))))...... ( -42.30) >DroVir_CAF1 26748 120 + 1 CGUUUCUGUGCCGGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUACGGCGACAUGGCCGUAUCGAACGCGAUUGGCUCAAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUUCC ............(((.(.(((((((.((.(((....(((((((((((((((((((......)))))((((....)))).))))).....)))))))))..))).)).))))))).).))) ( -41.60) >DroGri_CAF1 25335 120 + 1 CGUUUCUGUGCCGGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUUCGGCGACAUGGCCGUAUCGAAUGCAAUUGGCUCAAAUGUCUUUGAUAUACUAGUGUGCUUAGGACUUCC ............(((.(.(((((((.((.(((....(((((((((.(((....))....((((((.............)))).))..).)))))))))..))).)).))))))).).))) ( -37.22) >DroWil_CAF1 38235 120 + 1 CGUUUCUGUGCCUGAUGCCCUAAGCUCAAUAGCUGUUAUCAAAGAAGGCUUCGGCGAUAUGGCCGUAUCGAAUGCAAUUGGCUCGAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUUCC .............((.(.(((((((.((.(((.(((.((((((((......((((......))))..((((..((.....))))))...)))))))))))))).)).))))))).).)). ( -34.70) >DroMoj_CAF1 28280 120 + 1 CGUUUCUGUGCCUGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUACGGCGACAUGGCCGUUUCGAAUGCGAUUGGCUCAAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUUCC .............((.(.(((((((.((.(((....(((((((((((((((((((......))))).(((....)))..))))).....)))))))))..))).)).))))))).).)). ( -37.60) >DroAna_CAF1 24647 120 + 1 CGUGUCCGUCCCGGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUUCGGCGACAUGGCCGUAUCGAAUGCAAUUGGCUCGAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUCCC .(((((((...)))))))(((((((.((.(((....(((((((((.((((..(....)..))))...((((..((.....))))))...)))))))))..))).)).)))))))...... ( -42.30) >consensus CGUUUCUGUGCCGGAUGCCCUAAGCUCAAUAGCCGUUAUCAAAGAGGGCUUCGGCGACAUGGCCGUAUCGAAUGCAAUUGGCUCAAAUGUCUUUGAUAUACUAGUGUGCUUAGGUCUUCC .(..((((...))))..)(((((((.((.(((....((((((((((((((..(((......)))(((.....)))....))))).....)))))))))..))).)).)))))))...... (-34.22 = -34.80 + 0.58)

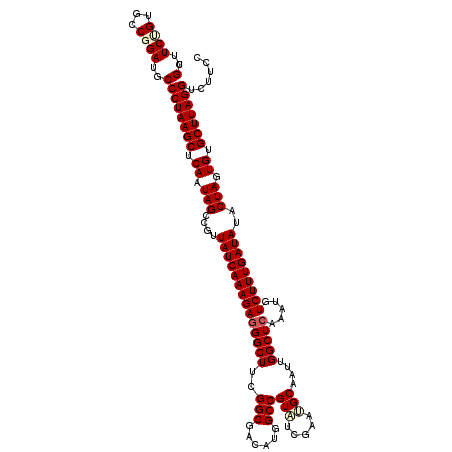
| Location | 191,084 – 191,204 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.94 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -35.46 |
| Energy contribution | -35.57 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.89 |
| SVM RNA-class probability | 0.997618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 191084 120 - 27905053 GGAAGACCUAAGCACACUAGUAUAUCAAAGACAUUCGAGCCUAUAGCGUUCGACACGGCCAUGUCGCCAAAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCCGGGACAGACACG (((.(.(((((((.((.((((.(((((((((...(((((((....).))))))...(((......))).......)))))))))...)))).)).))))))).).)))............ ( -40.90) >DroVir_CAF1 26748 120 - 1 GGAAGACCUAAGCACACUAGUAUAUCAAAGACAUUUGAGCCAAUCGCGUUCGAUACGGCCAUGUCGCCGUAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCCGGCACAGAAACG (((.(.(((((((.((.((((.(((((((((...(((((((....).))))))((((((......))))))....)))))))))...)))).)).))))))).).)))............ ( -43.20) >DroGri_CAF1 25335 120 - 1 GGAAGUCCUAAGCACACUAGUAUAUCAAAGACAUUUGAGCCAAUUGCAUUCGAUACGGCCAUGUCGCCGAAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCCGGCACAGAAACG (((.(((((((((.((.((((.(((((((((...((((((.....))..))))..((((......))))......)))))))))...)))).)).))))))))).)))............ ( -41.90) >DroWil_CAF1 38235 120 - 1 GGAAGACCUAAGCACACUAGUAUAUCAAAGACAUUCGAGCCAAUUGCAUUCGAUACGGCCAUAUCGCCGAAGCCUUCUUUGAUAACAGCUAUUGAGCUUAGGGCAUCAGGCACAGAAACG ......(((((((.((.((((.(((((((((...((((((.....))..))))..((((......))))......)))))))))...)))).)).)))))))((.....))......... ( -35.40) >DroMoj_CAF1 28280 120 - 1 GGAAGACCUAAGCACACUAGUAUAUCAAAGACAUUUGAGCCAAUCGCAUUCGAAACGGCCAUGUCGCCGUAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCAGGCACAGAAACG ......(((((((.((.((((.(((((((((..(((((((.....))..)))))(((((......))))).....)))))))))...)))).)).)))))))((.....))......... ( -36.70) >DroAna_CAF1 24647 120 - 1 GGGAGACCUAAGCACACUAGUAUAUCAAAGACAUUCGAGCCAAUUGCAUUCGAUACGGCCAUGUCGCCGAAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCCGGGACGGACACG .(((..(((((((.((.((((.(((((((((...((((((.....))..))))..((((......))))......)))))))))...)))).)).)))))))...)))............ ( -40.30) >consensus GGAAGACCUAAGCACACUAGUAUAUCAAAGACAUUCGAGCCAAUUGCAUUCGAUACGGCCAUGUCGCCGAAGCCCUCUUUGAUAACGGCUAUUGAGCUUAGGGCAUCCGGCACAGAAACG (((.(.(((((((.((.((((.(((((((((...((((((.....))..))))..((((......))))......)))))))))...)))).)).))))))).).)))............ (-35.46 = -35.57 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:26 2006