
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,978,373 – 5,978,504 |
| Length | 131 |
| Max. P | 0.964012 |

| Location | 5,978,373 – 5,978,466 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -30.52 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.75 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
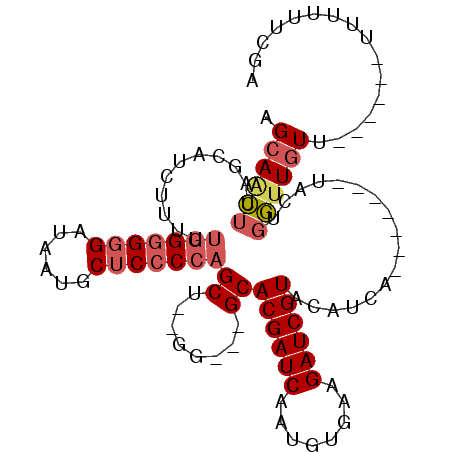
>3R_DroMel_CAF1 5978373 93 + 27905053 AGCAACUUGGCAUCUUUUUGGGGGGAUAAUGCUCCCCAGCU--GG---GCACGAUCAAUGUGAAGAUCGUACAUCA-------UGCUGGCUGUUU-U---UUUUUUCGA ((((..(..((((((..((((((((......))))))))..--))---(.((((((........)))))).)...)-------)))..).)))).-.---......... ( -27.50) >DroSec_CAF1 7065 101 + 1 AGCAACUUAGCAGCUUUUUGGGGGGAUAAUGCUCCCCAGCU--GG---GCACGAUCAAUGUGAAGAUCGUACAUCAUACAUCGUACUGGUUGUUUAU---UUUUUUCGA (((((((..((.((((.((((((((......))))))))..--))---))((((((........))))))............))...)))))))...---......... ( -31.00) >DroSim_CAF1 7128 97 + 1 AGCAACUUAGCAGCUUUUUGGGGGGAUAAUGCUCCCCAGCU--GG---GCACGAUCAAUUUGAAGAUCGUACAUCAUACAUCAUGCAGGUUGU-------UUUUUUCGA ((((((((.(((((((.((((((((......))))))))..--))---))((((((........)))))).............))))))))))-------)........ ( -34.50) >DroEre_CAF1 7786 89 + 1 AGCAACUCUGCAUCUUUUUGGGGGGAUAAUGCUCCCCAGCU--GG---GCACGAUCAAUGUGAAGAUCGUACAUCA-------UACUGGUUGUU--------UUUUCGA (((((((..((..((..((((((((......))))))))..--))---))((((((........))))))......-------....)))))))--------....... ( -27.00) >DroYak_CAF1 7514 91 + 1 AGCAACUUGGCAUCUUUUUGG-GGGAUAAUGCUCCCCAGCU--GGGGUGCACGAUCAGUGUGAGGAUCGUACAUCA-------UACUGGUUGUU--------UUUUCGA .(((.((..((..(.....)(-((((......))))).)).--.)).)))((((((((((((((.......).)))-------)))))))))).--------....... ( -35.10) >DroAna_CAF1 5973 85 + 1 AGAAGG---GC---------GGGGGAUAGUGC-CCCCAGCUCCGG---GCACGAUCAAUGUGAAGAUCGUACAUUA-------UGUUGCUUAUAC-UCCGUGUUUUCGA .((((.---((---------(((((((.((((-((........))---)))).)))((((((.......)))))).-------...........)-))))).))))... ( -28.00) >consensus AGCAACUUAGCAUCUUUUUGGGGGGAUAAUGCUCCCCAGCU__GG___GCACGAUCAAUGUGAAGAUCGUACAUCA_______UACUGGUUGUU______UUUUUUCGA .((((((...........(((((((......)))))))((........))((((((........)))))).................))))))................ (-18.97 = -19.75 + 0.78)

| Location | 5,978,373 – 5,978,466 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 79.13 |
| Mean single sequence MFE | -26.45 |
| Consensus MFE | -19.24 |
| Energy contribution | -19.43 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5978373 93 - 27905053 UCGAAAAAA---A-AAACAGCCAGCA-------UGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCAUUAUCCCCCCAAAAAGAUGCCAAGUUGCU .........---.-...((((..(((-------(...((((((((........))))))))---..--....(((((......)))))........))))...)))).. ( -26.40) >DroSec_CAF1 7065 101 - 1 UCGAAAAAA---AUAAACAACCAGUACGAUGUAUGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCAUUAUCCCCCCAAAAAGCUGCUAAGUUGCU .........---.....((((................((((((((........))))))))---.(--(((((((((......)))))......)))))....)))).. ( -27.20) >DroSim_CAF1 7128 97 - 1 UCGAAAAAA-------ACAACCUGCAUGAUGUAUGAUGUACGAUCUUCAAAUUGAUCGUGC---CC--AGCUGGGGAGCAUUAUCCCCCCAAAAAGCUGCUAAGUUGCU .........-------.((((..((............((((((((........))))))))---..--(((((((((......)))))......))))))...)))).. ( -27.40) >DroEre_CAF1 7786 89 - 1 UCGAAAA--------AACAACCAGUA-------UGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCAUUAUCCCCCCAAAAAGAUGCAGAGUUGCU .......--------..(((((.(((-------(...((((((((........))))))))---..--....(((((......)))))........)))).).)))).. ( -25.10) >DroYak_CAF1 7514 91 - 1 UCGAAAA--------AACAACCAGUA-------UGAUGUACGAUCCUCACACUGAUCGUGCACCCC--AGCUGGGGAGCAUUAUCCC-CCAAAAAGAUGCCAAGUUGCU .......--------..((((..(((-------(..(((((((((........)))))))))....--....(((((......))))-).......))))...)))).. ( -24.80) >DroAna_CAF1 5973 85 - 1 UCGAAAACACGGA-GUAUAAGCAACA-------UAAUGUACGAUCUUCACAUUGAUCGUGC---CCGGAGCUGGGG-GCACUAUCCCCC---------GC---CCUUCU ..(((.....((.-((....))....-------....((((((((........))))))))---))((.((.((((-(......)))))---------))---))))). ( -27.80) >consensus UCGAAAAAA______AACAACCAGCA_______UGAUGUACGAUCUUCACAUUGAUCGUGC___CC__AGCUGGGGAGCAUUAUCCCCCCAAAAAGAUGCCAAGUUGCU .................((((..((............((((((((........))))))))...........(((((......)))))..........))...)))).. (-19.24 = -19.43 + 0.20)



| Location | 5,978,402 – 5,978,504 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.04 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -14.58 |
| Energy contribution | -14.83 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5978402 102 - 27905053 --CAUAUCUCAAAGCUGGCAAUAAACGUGACAUAGAAGCCUCGAAAAAA---A-AAACAGCCAGCA-------UGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCA --...........((((((..................))).........---.-......(((((.-------((..((((((((........))))))))---.)--))))))..))). ( -26.67) >DroSec_CAF1 7094 93 - 1 ------------------UAAUAAACGUGACAAAGA-CCCUCGAAAAAA---AUAAACAACCAGUACGAUGUAUGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCA ------------------................(.-.(((((......---.....((..((......))..))..((((((((........))))))))---..--...)))))..). ( -17.40) >DroSim_CAF1 7157 104 - 1 --UAUCUC-CAAAGCUGGCAAUAAACGUGACAAAGA-CCCUCGAAAAAA-------ACAACCUGCAUGAUGUAUGAUGUACGAUCUUCAAAUUGAUCGUGC---CC--AGCUGGGGAGCA --...(((-(..((((((.......((((.((....-............-------......)))))).........((((((((........))))))))---))--))))..)))).. ( -29.95) >DroEre_CAF1 7815 98 - 1 UAUAUGUC-CGGAGCUGGCAAUAAACGUGACAAAGA-CUCUCGAAAA--------AACAACCAGUA-------UGAUGUACGAUCUUCACAUUGAUCGUGC---CC--AGCUGGGGAGCA ......((-(..((((((.......((.((......-.)).))....--------...........-------....((((((((........))))))))---))--))))..)))... ( -26.40) >DroYak_CAF1 7542 99 - 1 --UAUCUC-CAGAGCUGGCAAUAAACGUGACAAAGA-CACUCGAAAA--------AACAACCAGUA-------UGAUGUACGAUCCUCACACUGAUCGUGCACCCC--AGCUGGGGAGCA --...(((-(..((((((........(((.......-))).......--------...........-------...(((((((((........)))))))))..))--))))..)))).. ( -29.60) >DroAna_CAF1 5990 104 - 1 --CAUCUC-CGAAGCCGGCACUAAUCGUGACAACGA-CCCUCGAAAACACGGA-GUAUAAGCAACA-------UAAUGUACGAUCUUCACAUUGAUCGUGC---CCGGAGCUGGGG-GCA --..((((-((...((((.(((..(((((....((.-....))....))))))-))..........-------....((((((((........))))))))---))))...)))))-).. ( -30.60) >consensus __UAUCUC_CAAAGCUGGCAAUAAACGUGACAAAGA_CCCUCGAAAAAA______AACAACCAGCA_______UGAUGUACGAUCUUCACAUUGAUCGUGC___CC__AGCUGGGGAGCA ..........................((..(...((....))..................(((((............((((((((........))))))))........))))))..)). (-14.58 = -14.83 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:22 2006