
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,946,733 – 5,946,933 |
| Length | 200 |
| Max. P | 0.995173 |

| Location | 5,946,733 – 5,946,853 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -45.26 |
| Consensus MFE | -38.74 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946733 120 + 27905053 GGAUUCCGUCAUUUUGGCGGCUUUAUCGAGAGCAGCAGCUGCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGU (((((..((((....(((((.(((..((..((((((....))))))..))..)))))...((((((.......)))))))))....))))..))))).......(((((.....))))). ( -48.60) >DroSec_CAF1 59985 120 + 1 CGAUUCCGUCAUCUUGGCGGCUUUGUACAUAGCAGCCGCUGCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGU .((((..((((....(((((.(((((....((((((....))))))....)))))))...((((((.......)))))))))....))))..))))........(((((.....))))). ( -42.20) >DroSim_CAF1 48745 120 + 1 CGAUUCCGUCAUCUUGGCGGCUUUGUCCAUAGCAGCCGCUGCUGCUUCCGGUGAACCAUGUUGCUGGUAGAGACAGCAAGCCUUAUUGGCAUAUUCCUCCACUUCGGACAGCUUGUCCGU .......((((....(((((.((..((...((((((....))))))...))..))))...((((((.......)))))))))....))))..............(((((.....))))). ( -40.20) >DroEre_CAF1 50379 120 + 1 GGAUUCCGUCAUCUUGGCGGCUUUGUCCAGAGCAGCCGCUGCUGCUUCCGGCGAACCAUGCUGCUGGUAGAGACAGCAGGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGU (((((..((((....(((((.((((((..(((((((....)))))))..))))))))...((((((.......)))))))))....))))..))))).......(((((.....))))). ( -50.50) >DroYak_CAF1 52035 120 + 1 GGAUUCCGUCAUCUUGGCGGCUUUGUCCAGAGCAGCCGCUGCCGCUUCCGGUGAGCCGUGUUGCUGGUAGAGACAGCAGGCCCUAUUGGCAUAGUUCUCCACUUCGGGCAGCUGGUCGGU .....((((((....(((.(((((....))))).)))((((((......((.((((..(.((((((.......))))))(((.....))))..)))).))......))))))))).))). ( -44.80) >consensus GGAUUCCGUCAUCUUGGCGGCUUUGUCCAGAGCAGCCGCUGCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGU (((((..((((....(((((.((((((..(((((((....)))))))..))))))))...((((((.......)))))))))....))))..))))).......(((((.....))))). (-38.74 = -39.50 + 0.76)

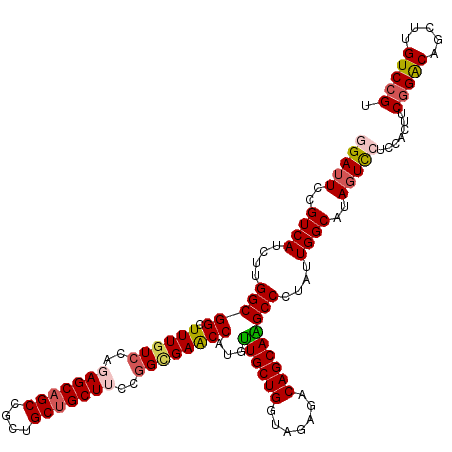

| Location | 5,946,733 – 5,946,853 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -42.86 |
| Consensus MFE | -35.82 |
| Energy contribution | -37.26 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946733 120 - 27905053 ACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGCAGCUGCUGCUCUCGAUAAAGCCGCCAAAAUGACGGAAUCC .(((((.....)))))..((((.((((((((((.....)))((((((.......)))))).)))))))..((..((((((....))))))..))......))))................ ( -43.70) >DroSec_CAF1 59985 120 - 1 ACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGCAGCGGCUGCUAUGUACAAAGCCGCCAAGAUGACGGAAUCG .(((((.....)))))..((((.((((((((((.....)))((((((.......)))))).)))))))......((((((....))))))..........))))...(((......))). ( -40.30) >DroSim_CAF1 48745 120 - 1 ACGGACAAGCUGUCCGAAGUGGAGGAAUAUGCCAAUAAGGCUUGCUGUCUCUACCAGCAACAUGGUUCACCGGAAGCAGCAGCGGCUGCUAUGGACAAAGCCGCCAAGAUGACGGAAUCG .(((((.....)))))..((((..((((..(((.....)))((((((.......))))))....)))).(((..((((((....)))))).)))......))))...(((......))). ( -39.10) >DroEre_CAF1 50379 120 - 1 ACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCCUGCUGUCUCUACCAGCAGCAUGGUUCGCCGGAAGCAGCAGCGGCUGCUCUGGACAAAGCCGCCAAGAUGACGGAAUCC .(((((.....)))))..((((.((((((((((.....)))((((((.......)))))).)))))))((((((.(((((....)))))))))).)....))))...(((......))). ( -47.30) >DroYak_CAF1 52035 120 - 1 ACCGACCAGCUGCCCGAAGUGGAGAACUAUGCCAAUAGGGCCUGCUGUCUCUACCAGCAACACGGCUCACCGGAAGCGGCAGCGGCUGCUCUGGACAAAGCCGCCAAGAUGACGGAAUCC .(((....((((((....(((((((..((.(((.....))).))...)))))))..((..(.(((....))))..))))))))(((.(((........))).))).......)))..... ( -43.90) >consensus ACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGCAGCGGCUGCUCUGGACAAAGCCGCCAAGAUGACGGAAUCC .(((((.....)))))..((((.((((((((((.....)))((((((.......)))))).))))))).(((((.(((((....))))))))))......))))................ (-35.82 = -37.26 + 1.44)

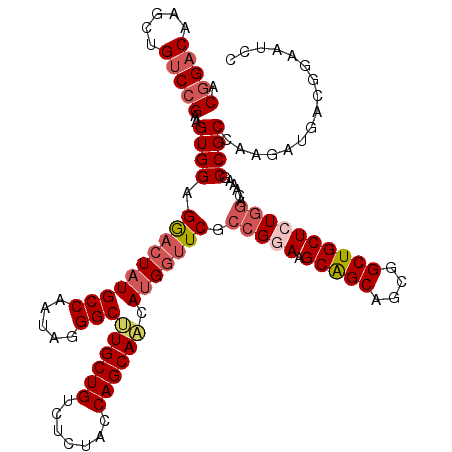

| Location | 5,946,773 – 5,946,893 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -39.80 |
| Energy contribution | -39.24 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946773 120 + 27905053 GCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCU ((((..(((.((((......((((..(.((((((.((((((((....))....((((........)))).))))))..)))))))..))))((((......)))))))).)))..)))). ( -42.50) >DroSec_CAF1 60025 120 + 1 GCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCU ((((..(((.((((......((((..(.((((((.((((((((....))....((((........)))).))))))..)))))))..))))((((......)))))))).)))..)))). ( -42.50) >DroSim_CAF1 48785 120 + 1 GCUGCUUCCGGUGAACCAUGUUGCUGGUAGAGACAGCAAGCCUUAUUGGCAUAUUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCU ((((..(((((....))...((((..(.((((((.....(((.....)))...............((((.....)))))))))))..))))((((......)))).....)))..)))). ( -40.50) >DroEre_CAF1 50419 120 + 1 GCUGCUUCCGGCGAACCAUGCUGCUGGUAGAGACAGCAGGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCU ((((..(((.((((.......(((..(.((((((.((((((((....))....((((........)))).))))))..)))))))..))).((((......)))))))).)))..)))). ( -43.60) >DroYak_CAF1 52075 120 + 1 GCCGCUUCCGGUGAGCCGUGUUGCUGGUAGAGACAGCAGGCCCUAUUGGCAUAGUUCUCCACUUCGGGCAGCUGGUCGGUUUCUUUGGCCAGCAGGAUAAUCUGCUCGUAGGACUUGGCU ((((..(((((.((((..(.((((((.......))))))(((.....))))..)))).))....(((((.(((((((((.....))))))))).((.....)))))))..)))..)))). ( -49.00) >consensus GCUGCUUCCGGCGAACCAUGUUGCUGGUAGAGACAGCAAGCCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCU ((((..(((..(((......((((..(.((((((.((((((((....))....((((........)))).))))))..)))))))..))))((((......)))))))..)))..)))). (-39.80 = -39.24 + -0.56)



| Location | 5,946,773 – 5,946,893 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.50 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -35.04 |
| Energy contribution | -36.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946773 120 - 27905053 AGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGC ((((...((((.(((((((......)))))))(((......(((((.....)))))...)))))))((((....)))))))).((((..(((((((......)))).....)))..)))) ( -42.40) >DroSec_CAF1 60025 120 - 1 AGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGC ((((...((((.(((((((......)))))))(((......(((((.....)))))...)))))))((((....)))))))).((((..(((((((......)))).....)))..)))) ( -42.40) >DroSim_CAF1 48785 120 - 1 AGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGAAUAUGCCAAUAAGGCUUGCUGUCUCUACCAGCAACAUGGUUCACCGGAAGCAGC ((((...((((.(((((((......)))))))(((......(((((.....)))))...))))))).(((....))).)))).((((..(((((((......)))).....)))..)))) ( -39.60) >DroEre_CAF1 50419 120 - 1 AGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCCUGCUGUCUCUACCAGCAGCAUGGUUCGCCGGAAGCAGC .((.........(((((((......)))))))((.......(((((.....)))))....((.((((((((((.....)))((((((.......)))))).))))))).))))..))... ( -42.30) >DroYak_CAF1 52075 120 - 1 AGCCAAGUCCUACGAGCAGAUUAUCCUGCUGGCCAAAGAAACCGACCAGCUGCCCGAAGUGGAGAACUAUGCCAAUAGGGCCUGCUGUCUCUACCAGCAACACGGCUCACCGGAAGCGGC .(((...(((...((((.........((((((....(((....(((.(((.((((....(((.........)))...))))..))))))))).)))))).....))))...)))...))) ( -38.04) >consensus AGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGGCUUGCUGUCUCUACCAGCAACAUGGUUCGCCGGAAGCAGC .((....(((..(((((((......))))))).........(((((.....)))))....((.((((((((((.....)))((((((.......)))))).))))))).))))).))... (-35.04 = -36.04 + 1.00)


| Location | 5,946,813 – 5,946,933 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -34.94 |
| Energy contribution | -34.14 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946813 120 + 27905053 CCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCUGCAUGGAACCAUGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((..(((((((.....)))..(((((.(((......)))))))).))))....)))).)))))..(((((((.((((((.((........)).))))))..)))))))... ( -35.70) >DroSec_CAF1 60065 120 + 1 CCCUAUUGGCAUAGUCCUCCACUUCGGACAGUUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCUGCAUGGAACCAUGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((..(((((((.....)))..(((((.(((......)))))))).))))....)))).)))))..(((((((.((((((.((........)).))))))..)))))))... ( -35.70) >DroSim_CAF1 48825 120 + 1 CCUUAUUGGCAUAUUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCUGCAUGGAACCAUGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((.(((..(((.(....)...(((((.(((......)))))))).))).))).)))).)))))..(((((((.((((((.((........)).))))))..)))))))... ( -34.60) >DroEre_CAF1 50459 120 + 1 CCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCUGCAUGGAACCACGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((..(((((((.....)))..(((((.(((......)))))))).))))....)))).)))))..(((((((.((((((..((((....))))))))))..)))))))... ( -38.60) >DroYak_CAF1 52115 120 + 1 CCCUAUUGGCAUAGUUCUCCACUUCGGGCAGCUGGUCGGUUUCUUUGGCCAGCAGGAUAAUCUGCUCGUAGGACUUGGCUGCGUGGAACCACGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((..(((.(((.(....)...(((((((((.....))))))))).))).))).)))).)))))..(((((((((((....))))((.....))........)))))))... ( -41.50) >consensus CCCUAUUGGCAUAGUCCUCCACUUCGGACAGCUUGUCCGUUUCUUUGGCAAGCAGGAUAAUCUGCUCGUAGGAUUUGGCUGCAUGGAACCAUGACUUGUUGUUUUCGUAACAGUCGAUGG .(((((.((((..(((((((.....)))..(((((.(((......)))))))).))))....)))).)))))..(((((((.((((((.((........)).))))))..)))))))... (-34.94 = -34.14 + -0.80)

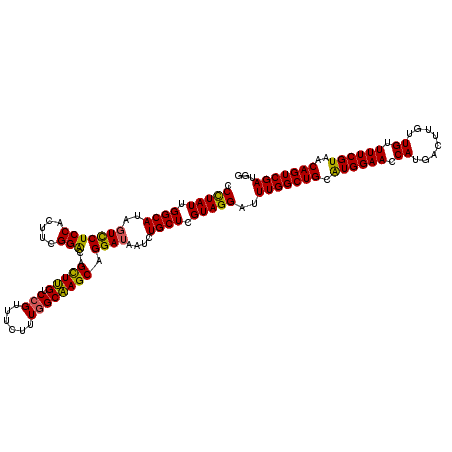
| Location | 5,946,813 – 5,946,933 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.00 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -30.20 |
| Energy contribution | -30.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5946813 120 - 27905053 CCAUCGACUGUUACGAAAACAACAAGUCAUGGUUCCAUGCAGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGG ((((.(((((((........))).))))))))..........((...((((.(((((((......)))))))(((......(((((.....)))))...)))))))((((....)))))) ( -34.10) >DroSec_CAF1 60065 120 - 1 CCAUCGACUGUUACGAAAACAACAAGUCAUGGUUCCAUGCAGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAACUGUCCGAAGUGGAGGACUAUGCCAAUAGGG ((((.(((((((........))).))))))))..........((...((((.(((((((......)))))))(((......(((((.....)))))...)))))))((((....)))))) ( -34.10) >DroSim_CAF1 48825 120 - 1 CCAUCGACUGUUACGAAAACAACAAGUCAUGGUUCCAUGCAGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGAAUAUGCCAAUAAGG ((((.(((((((........))).))))))))......(((......((((.(((((((......)))))))(((......(((((.....)))))...)))))))...)))........ ( -33.20) >DroEre_CAF1 50459 120 - 1 CCAUCGACUGUUACGAAAACAACAAGUCGUGGUUCCAUGCAGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGG (((.((((((((........))).))))))))..........((...((((.(((((((......)))))))(((......(((((.....)))))...)))))))((((....)))))) ( -35.50) >DroYak_CAF1 52115 120 - 1 CCAUCGACUGUUACGAAAACAACAAGUCGUGGUUCCACGCAGCCAAGUCCUACGAGCAGAUUAUCCUGCUGGCCAAAGAAACCGACCAGCUGCCCGAAGUGGAGAACUAUGCCAAUAGGG (((.((((((((........))).))))))))(((((((((((...(((.....(((((......)))))((.........)))))..))))).....))))))..((((....)))).. ( -34.70) >consensus CCAUCGACUGUUACGAAAACAACAAGUCAUGGUUCCAUGCAGCCAAAUCCUACGAGCAGAUUAUCCUGCUUGCCAAAGAAACGGACAAGCUGUCCGAAGUGGAGGACUAUGCCAAUAGGG ((((.(((((((........))).))))))))((((((..............(((((((......))))))).........(((((.....)))))..))))))..((((....)))).. (-30.20 = -30.60 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:08 2006