
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,939,481 – 5,939,581 |
| Length | 100 |
| Max. P | 0.787384 |

| Location | 5,939,481 – 5,939,581 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 69.49 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -5.76 |
| Energy contribution | -6.98 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.21 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5939481 100 + 27905053 AUGUGAUAAGGUCAGCAUAAGCUAAAUAUUUGGUCACGCUGGCGAGCGGACUUGUGAGUUCGGAAAACUCACA-CUGCUGACCAC--UGCUGAUCGAGAUAAU .........((((((((..............((((.((((....))))))))((((((((.....))))))))-.))))))))..--................ ( -32.90) >DroPse_CAF1 56125 78 + 1 -UGCGAUGGGGUCACCGAAUGUUAAAAAUUUCGUAA-------------AUUUAUAGCUGCGGGGA---AAAA-CUGCUGAC-------CCAAUCAAGAUAAU -...(((.((((((......((((.(((((.....)-------------)))).)))).((((...---....-))))))))-------)).)))........ ( -20.20) >DroSec_CAF1 52891 102 + 1 AUGUGAUAAGGUCAGCAGAAGCUAAAUAUUUGGUCACGCUGGGUAGCGGACUUGUGAGUUCGGAGAACUCACA-CUGCUGACCAUCCAUCCAAUCGAGAUAAU (((.(((..(((((((((.............((((.((((....))))))))(((((((((...)))))))))-))))))))))))))).............. ( -38.60) >DroSim_CAF1 41634 103 + 1 AUGUGAUAAGGUCAGCAGAAGCUAAAUAUUUGGUCACGCUGGGUAGCGGACUUGUGAGUUCGGAAAACUCACAAACGCUGACCAUCCAUCCAAUCGAGAUAAU (((.(((..(((((((................(((.((((....)))))))(((((((((.....)))))))))..))))))))))))).............. ( -33.10) >DroEre_CAF1 43340 89 + 1 AUGUGAUAAGGUCAGCAGAAGCCAAAUAGCUGGUCACGGUGGGCAGUGGACUUGUAGGAUCGGAAACC-AAAA-CU------------GCCGAUGGAGAUAAU .........(..((((............))))..).(.((.((((((.((((....)).))(....).-...)-))------------))).)).)....... ( -22.90) >DroYak_CAF1 44725 82 + 1 AUGUGAUAAGGUCAGCAGAAGCUAAGUAUCUGGUCACGAUGGCCAGUGGACUUGUGGGAUCGGAAAAC---------------------CCAAUAGAGAUAAU .........(((((((....)))((((..((((((.....))))))...))))....))))((.....---------------------))............ ( -19.50) >consensus AUGUGAUAAGGUCAGCAGAAGCUAAAUAUUUGGUCACGCUGGGCAGCGGACUUGUGAGUUCGGAAAAC_CACA_CUGCUGAC_______CCAAUCGAGAUAAU ..(((((.((((.(((....)))....)))).))))).........((((((....))))))......................................... ( -5.76 = -6.98 + 1.22)

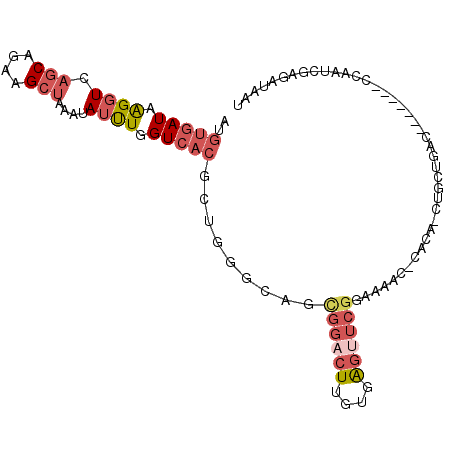

| Location | 5,939,481 – 5,939,581 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 69.49 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -4.25 |
| Energy contribution | -4.47 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.17 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5939481 100 - 27905053 AUUAUCUCGAUCAGCA--GUGGUCAGCAG-UGUGAGUUUUCCGAACUCACAAGUCCGCUCGCCAGCGUGACCAAAUAUUUAGCUUAUGCUGACCUUAUCACAU ................--..((((((((.-(((((((((...))))))))).(((((((....)))).)))...............))))))))......... ( -31.90) >DroPse_CAF1 56125 78 - 1 AUUAUCUUGAUUGG-------GUCAGCAG-UUUU---UCCCCGCAGCUAUAAAU-------------UUACGAAAUUUUUAACAUUCGGUGACCCCAUCGCA- ........(((.((-------((((((.(-(...---.....)).)))......-------------...((((..........))))..))))).)))...- ( -15.60) >DroSec_CAF1 52891 102 - 1 AUUAUCUCGAUUGGAUGGAUGGUCAGCAG-UGUGAGUUCUCCGAACUCACAAGUCCGCUACCCAGCGUGACCAAAUAUUUAGCUUCUGCUGACCUUAUCACAU ...........((((((...(((((((((-(((((((((...))))))))).(((((((....)))).)))..............))))))))).)))).)). ( -38.40) >DroSim_CAF1 41634 103 - 1 AUUAUCUCGAUUGGAUGGAUGGUCAGCGUUUGUGAGUUUUCCGAACUCACAAGUCCGCUACCCAGCGUGACCAAAUAUUUAGCUUCUGCUGACCUUAUCACAU ...........((((((...(((((((..((((((((((...))))))))))(((((((....)))).)))................))))))).)))).)). ( -34.50) >DroEre_CAF1 43340 89 - 1 AUUAUCUCCAUCGGC------------AG-UUUU-GGUUUCCGAUCCUACAAGUCCACUGCCCACCGUGACCAGCUAUUUGGCUUCUGCUGACCUUAUCACAU ............(((------------((-((((-(.............))))...))))))....((((..((....(..((....))..).))..)))).. ( -16.82) >DroYak_CAF1 44725 82 - 1 AUUAUCUCUAUUGG---------------------GUUUUCCGAUCCCACAAGUCCACUGGCCAUCGUGACCAGAUACUUAGCUUCUGCUGACCUUAUCACAU .............(---------------------(((...((((.(((.........)))..)))).)))).((((.(((((....)))))...)))).... ( -14.40) >consensus AUUAUCUCGAUUGG_______GUCAGCAG_UGUG_GUUUUCCGAACCCACAAGUCCGCUACCCAGCGUGACCAAAUAUUUAGCUUCUGCUGACCUUAUCACAU ..................................................................((((........(((((....))))).....)))).. ( -4.25 = -4.47 + 0.22)

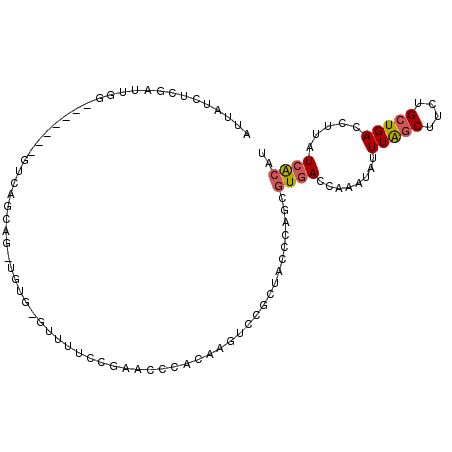
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:27:00 2006