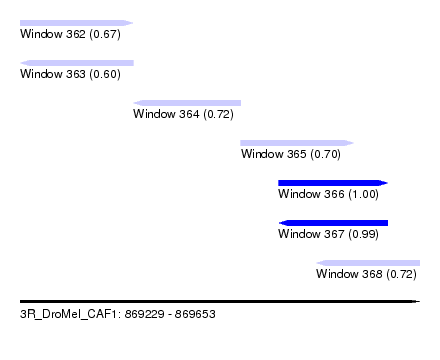
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 869,229 – 869,653 |
| Length | 424 |
| Max. P | 0.995236 |

| Location | 869,229 – 869,349 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -35.58 |
| Consensus MFE | -31.17 |
| Energy contribution | -31.93 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869229 120 + 27905053 UUUAGCUACAGAUAUGGCCCAGCAGUCGGCCCAUAAGUCGCGUUAUUUGUAUUUGUGCCAGUAUCUGUGGCCGUAUCCGCAUCCGCAUCCGUAGCUGUAGCUGUAGCUGUAUCUGUAUCU ..(((((((.((((((((((((..(..(((..((((((.(((.....))))))))))))..)..))).))))))))).((....))....)))))))((((....))))........... ( -39.10) >DroSec_CAF1 44251 113 + 1 UUUACCUACAGAUAUGGCCCAGAAGUCGGCCCAUAAGUCGCGUUAUUUGUAUUUGUGCCAGUAUCUGUGGCCGUAUCCGC------AUCCGUAGCUGUGGCUCUAGCUGUAUCUGUAUC- ......(((((((((((((((((.(..(((..((((((.(((.....))))))))))))..).)))).))))....((((------(........)))))........)))))))))..- ( -35.80) >DroSim_CAF1 43456 113 + 1 UUUAGCUACAGAUAUGGCCCAGAAGUCGGUCCAUAAGUCGCGUUAUUUGUAUUUGUGCCAGUAUCCGUGGCCGUAUCCGC------AUCCGUAGCUGUAGCUCUAGCUGUAUCUGUAUC- ..(((((((.((((((((((.((.(..((..(((((((.(((.....))))))))))))..).)).).))))))))).(.------...))))))))((((....))))..........- ( -32.70) >DroEre_CAF1 43469 113 + 1 UCUAGCUACAGAUAUGACCCAGAAGACGGCCCAUAAGUCGCGUUAUUUGUGCUUGUGCCAGUAUCUGUGGCUGUAUCCGU------AUCCGUAGCUGAAGCUGUAGCUGUAGCCGUAUC- ....(((((((((((..((((((..(((((..(((((.((((.....)))))))))))).)).)))).))..)))))...------..(..((((....))))..).))))))......- ( -34.70) >consensus UUUAGCUACAGAUAUGGCCCAGAAGUCGGCCCAUAAGUCGCGUUAUUUGUAUUUGUGCCAGUAUCUGUGGCCGUAUCCGC______AUCCGUAGCUGUAGCUCUAGCUGUAUCUGUAUC_ ..(((((((((((((((((((((.(..(((..((((((.(((.....))))))))))))..).)))).)))))))))................((....))))))))))........... (-31.17 = -31.93 + 0.75)

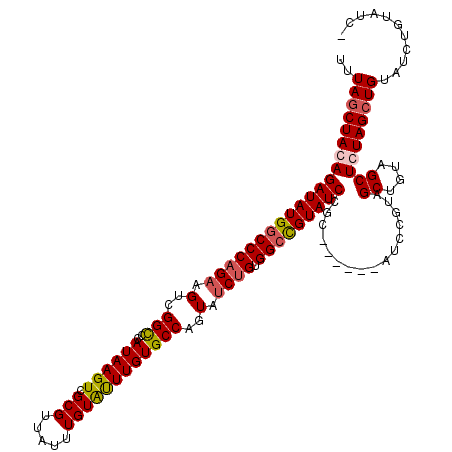
| Location | 869,229 – 869,349 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.99 |
| Mean single sequence MFE | -32.84 |
| Consensus MFE | -27.14 |
| Energy contribution | -27.57 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.602241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869229 120 - 27905053 AGAUACAGAUACAGCUACAGCUACAGCUACGGAUGCGGAUGCGGAUACGGCCACAGAUACUGGCACAAAUACAAAUAACGCGACUUAUGGGCCGACUGCUGGGCCAUAUCUGUAGCUAAA ............(((....)))..(((((((((((.((..((((...(((((.((.....(.((...............)).)....))))))).))))....)).)))))))))))... ( -36.66) >DroSec_CAF1 44251 113 - 1 -GAUACAGAUACAGCUAGAGCCACAGCUACGGAU------GCGGAUACGGCCACAGAUACUGGCACAAAUACAAAUAACGCGACUUAUGGGCCGACUUCUGGGCCAUAUCUGUAGGUAAA -..((((((((.........((.((.(....).)------).))....((((.((((...((((.((....................)).))))...)))))))).))))))))...... ( -31.75) >DroSim_CAF1 43456 113 - 1 -GAUACAGAUACAGCUAGAGCUACAGCUACGGAU------GCGGAUACGGCCACGGAUACUGGCACAAAUACAAAUAACGCGACUUAUGGACCGACUUCUGGGCCAUAUCUGUAGCUAAA -...........(((....)))..((((((((((------(.(....)((((.((((...(((..((....................))..)))...)))))))).)))))))))))... ( -31.55) >DroEre_CAF1 43469 113 - 1 -GAUACGGCUACAGCUACAGCUUCAGCUACGGAU------ACGGAUACAGCCACAGAUACUGGCACAAGCACAAAUAACGCGACUUAUGGGCCGUCUUCUGGGUCAUAUCUGUAGCUAGA -.....((((..(((....)))..))))..((.(------(((((((..(((.((((...((((..((((.(.......).).)))....))))...)))))))..)))))))).))... ( -31.40) >consensus _GAUACAGAUACAGCUACAGCUACAGCUACGGAU______GCGGAUACGGCCACAGAUACUGGCACAAAUACAAAUAACGCGACUUAUGGGCCGACUUCUGGGCCAUAUCUGUAGCUAAA ............((((..(((....)))............(((((((.((((.((((...((((.((....................)).))))...)))))))).)))))))))))... (-27.14 = -27.57 + 0.44)

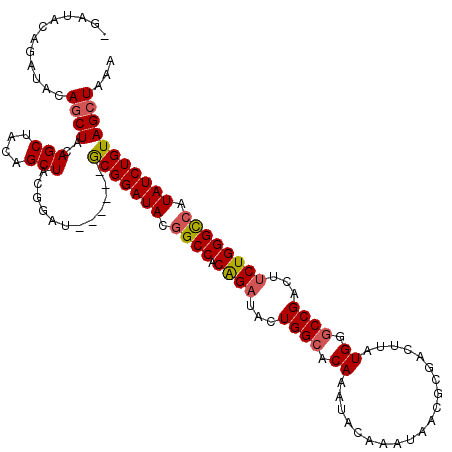
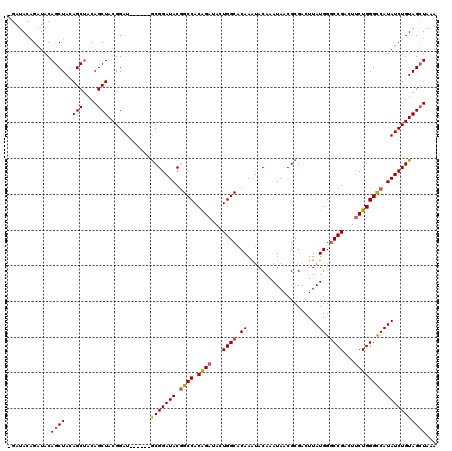
| Location | 869,349 – 869,463 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -30.67 |
| Energy contribution | -30.30 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869349 114 - 27905053 AGCCUUGGAUUAGUCGCAGUCAGCUGCAUCUGACGGAUGCAGAUACUCGCACU------CGCACUCCCUUUUGUGCAAGCACCAAAAAAAAACUGGCACAAAUGUCACGCAGCCAUAUAC .((.((((...(((.(((((...(((((((.....)))))))..))).)))))------.((((........)))).....))))........(((((....))))).)).......... ( -30.30) >DroSec_CAF1 44364 108 - 1 UGCCUUGGAUUAGUCGCAGUCAGCUGCAUCUGACGGAUGCAGAUACUCGCACU------CGCACUCGCUGUUGUGCAAGAACCGAAA-AAAACUGGCACAAAUGUCACGCAGCCA----- (((.((((.......(((((...(((((((.....)))))))..))).)).((------.((((........)))).))..))))..-.....(((((....))))).)))....----- ( -30.70) >DroSim_CAF1 43569 108 - 1 GGCCUCGGAUUAGUCGCAGUCAGCUGCAUCUGACGGAUGCAGAUACUCGCACU------UGCACUCGCUGUUGUGCAAGAACCGAAA-AAAACUGGCACAAAUGUCACGCAGCCA----- (((.((((.......(((((...(((((((.....)))))))..))).)).((------(((((........)))))))..))))..-.....(((((....)))))....))).----- ( -39.40) >DroEre_CAF1 43582 115 - 1 UGCCUUGGAUUAGUCGCAGUCAGCUGCAUCUGACGGAUGCAGAUACUCGCACUCGCACUCGCACUCGCUGUUGUGCAAGCACCGAAAAAAAACUGGCACAAAUGUCACGCAGCCA----- (((.((((...(((.(((((...(((((((.....)))))))..))).))))).((((..((....))....)))).....))))........(((((....))))).)))....----- ( -31.10) >consensus UGCCUUGGAUUAGUCGCAGUCAGCUGCAUCUGACGGAUGCAGAUACUCGCACU______CGCACUCGCUGUUGUGCAAGAACCGAAA_AAAACUGGCACAAAUGUCACGCAGCCA_____ .((.((((...(((.(((((...(((((((.....)))))))..))).))))).......((((........)))).....))))........(((((....))))).)).......... (-30.67 = -30.30 + -0.37)

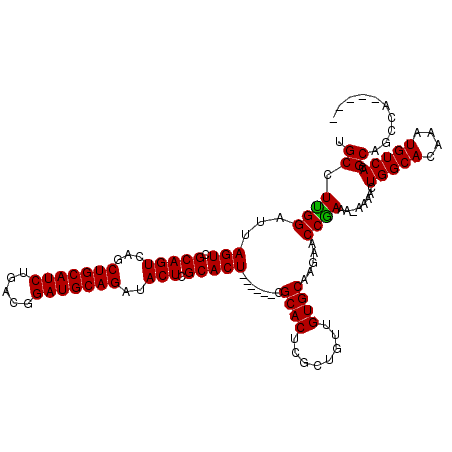
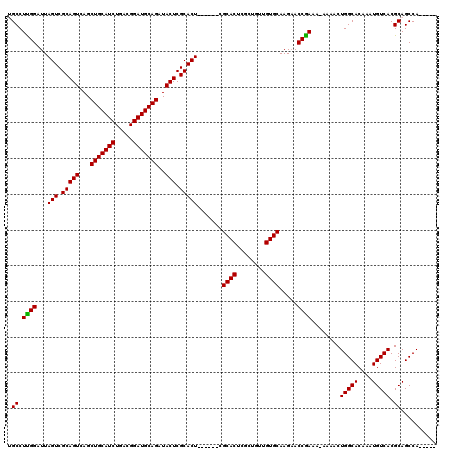
| Location | 869,463 – 869,583 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -20.73 |
| Energy contribution | -21.35 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700813 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869463 120 + 27905053 GAAUUCGGCGGGGUUCCUGGCUUACACUCAGCCACUUCGCAUUUUAAUUAAAACGAAUUGCGCAUUAUUUGCGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUAAG .......(((((((....((((.......))))))))))).((((((((((........(((((.....)))))..(((.((((..........)))))))))))))))))......... ( -29.00) >DroSec_CAF1 44472 120 + 1 CAAUUCGGCGGGGUUCCUGCCUUACACUCAGCCACUUCGCAUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAG ......((((((...)))))).........((((.((((..((......))..))))..(((((.....))))).)))).......(((((((((......))))))))).......... ( -25.40) >DroSim_CAF1 43677 120 + 1 CAAUUCGGCGGGGUUCCUGCCUUACACUCAGCCACUUCGCAUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAG ......((((((...)))))).........((((.((((..((......))..))))..(((((.....))))).)))).......(((((((((......))))))))).......... ( -25.40) >DroEre_CAF1 43697 120 + 1 CCAUUCAGGGGGGGUCCUACCUUACAUUCCGCCACUUCGCAUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUUUAAUUUAAUUAAUAAGCCCUAAUUAAAACUUAAUUAG .......((.(((((..........))))).)).(((........((((((((((....(((((.....)))))....))))))))))........)))..(((((((.....))))))) ( -25.09) >consensus CAAUUCGGCGGGGUUCCUGCCUUACACUCAGCCACUUCGCAUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAG ......((((((...)))))).........((((.((((..((......))..))))..(((((.....))))).)))).......(((((((((......))))))))).......... (-20.73 = -21.35 + 0.62)

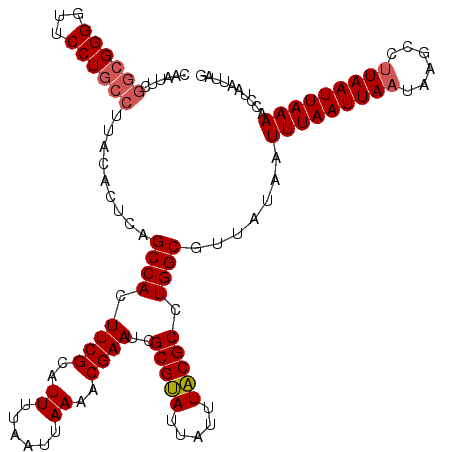
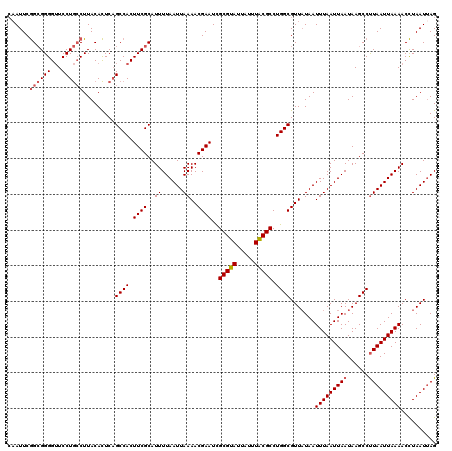
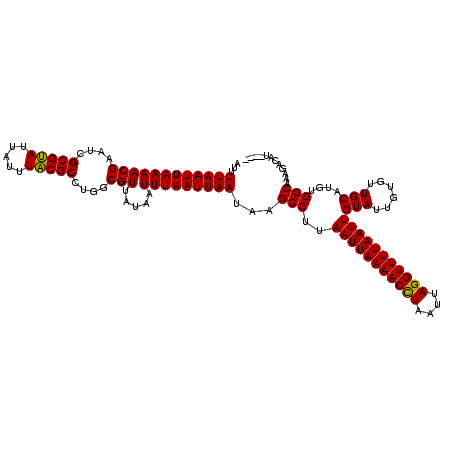
| Location | 869,503 – 869,619 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -27.89 |
| Energy contribution | -27.32 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869503 116 + 27905053 AUUUUAAUUAAAACGAAUUGCGCAUUAUUUGCGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUAAGGUUUUAAUUGUAUUGUGUUGCAUGUGGCAAAGACAU---- ...((((((((((((....(((((.....)))))....))).....)))))))))...(((..(((((((((((....)))))))))))(((......)))....)))........---- ( -29.60) >DroSec_CAF1 44512 120 + 1 AUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAGGUUUUAAUUGUAUUGUGUUGCAUGUGGCAAAGACAUUCCG ...((((((((((((....(((((.....)))))....))).....)))))))))...(((..(((((((((((....)))))))))))(((......)))....)))............ ( -27.60) >DroSim_CAF1 43717 116 + 1 AUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAGGUUUUAAUUGUAUUGUGUUGCAUGUGGCAAAGACAU---- ...((((((((((((....(((((.....)))))....))).....)))))))))...(((..(((((((((((....)))))))))))(((......)))....)))........---- ( -27.60) >DroEre_CAF1 43737 116 + 1 AUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUUUAAUUUAAUUAAUAAGCCCUAAUUAAAACUUAAUUAGGUUUUAAUUGUAUUGUGUUGCAUGUGGCAAAGACAU---- .....((((((((((....(((((.....)))))....))))))))))..........(((..(((((((((((....)))))))))))(((......)))....)))........---- ( -27.70) >consensus AUUUUAAUUAAAACGAAUCGCGUAUUAUUUACGCCUGGCGUUAUAAUUUAAUUAAUAAGCCUUAAUUAAAACCUAAUUAGGUUUUAAUUGUAUUGUGUUGCAUGUGGCAAAGACAU____ ...((((((((((((....(((((.....)))))....))).....)))))))))...(((..(((((((((((....)))))))))))(((......)))....)))............ (-27.89 = -27.32 + -0.56)

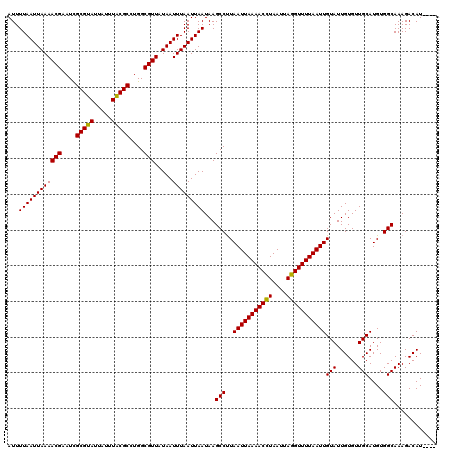
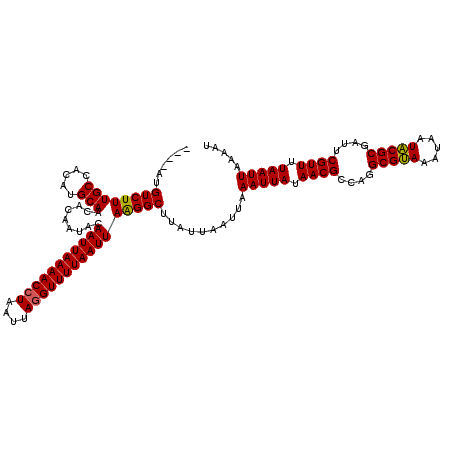
| Location | 869,503 – 869,619 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.34 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -24.59 |
| Energy contribution | -24.28 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869503 116 - 27905053 ----AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUUAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAACGCCAGGCGCAAAUAAUGCGCAAUUCGUUUUAAUUAAAAU ----..((((((((.....))).........(((((((((((....))))))))))))))))..........(((((.((((....(((((.....)))))....)))).)))))..... ( -28.30) >DroSec_CAF1 44512 120 - 1 CGGAAUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAACGCCAGGCGUAAAUAAUACGCGAUUCGUUUUAAUUAAAAU ......((((((((.....))).........(((((((((((....))))))))))))))))..........(((((.((((....(((((.....)))))....)))).)))))..... ( -24.90) >DroSim_CAF1 43717 116 - 1 ----AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAACGCCAGGCGUAAAUAAUACGCGAUUCGUUUUAAUUAAAAU ----..((((((((.....))).........(((((((((((....))))))))))))))))..........(((((.((((....(((((.....)))))....)))).)))))..... ( -24.90) >DroEre_CAF1 43737 116 - 1 ----AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAAGUUUUAAUUAGGGCUUAUUAAUUAAAUUAAAACGCCAGGCGUAAAUAAUACGCGAUUCGUUUUAAUUAAAAU ----.(((..((((.....)))).))).......(((..((((((((.....))))))))..))).......((((((((((....(((((.....)))))....))))))))))..... ( -24.60) >consensus ____AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAACGCCAGGCGUAAAUAAUACGCGAUUCGUUUUAAUUAAAAU ......((((((((.....))).........(((((((((((....))))))))))))))))..........(((((.((((....(((((.....)))))....)))).)))))..... (-24.59 = -24.28 + -0.31)

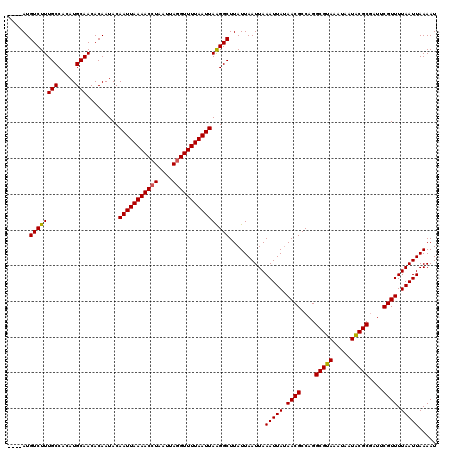
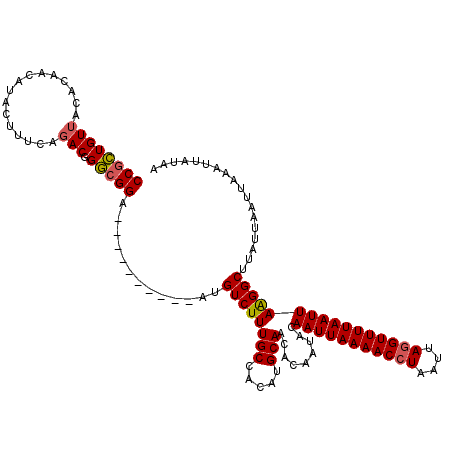
| Location | 869,543 – 869,653 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -21.55 |
| Consensus MFE | -17.10 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 869543 110 - 27905053 CCGCUGUUACAAAACAUACUUUCAGACGGGCGGA----------AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUUAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAA ((((((((..(((......)))..))).))))).----------..((((((((.....))).........(((((((((((....)))))))))))))))).................. ( -21.70) >DroSec_CAF1 44552 120 - 1 CCGCUGUUACACAACAUACUUUCAGACGGGCGGAAUGUCGCGGAAUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAA ((((((((....))))........((((.......))))))))...((((((((.....))).........(((((((((((....)))))))))))))))).................. ( -24.60) >DroSim_CAF1 43757 110 - 1 CCGCUGUUACACAACAUACUUUCAGACGGGCGGA----------AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAA ((((((((................))).))))).----------..((((((((.....))).........(((((((((((....)))))))))))))))).................. ( -21.39) >DroEre_CAF1 43777 110 - 1 CCAGUGUAGCGCAAUAUAUUUUCAGACGGUCGGA----------AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAAGUUUUAAUUAGGGCUUAUUAAUUAAAUUAAAA ...((((.((((((.........(((((......----------.))))))))).....)).))))........(((..((((((((.....))))))))..)))............... ( -18.50) >consensus CCGCUGUUACACAACAUACUUUCAGACGGGCGGA__________AUGUCUUUGCCACAUGCAACACAAUACAAUUAAAACCUAAUUAGGUUUUAAUUAAGGCUUAUUAAUUAAAUUAUAA ((((((((................))).))))).............((((((((.....))).........(((((((((((....)))))))))))))))).................. (-17.10 = -17.29 + 0.19)

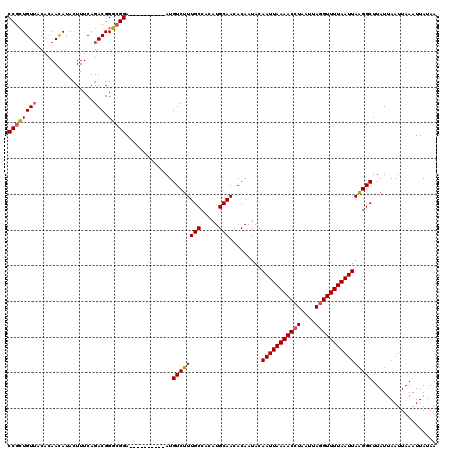
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:32 2006