
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,882,776 – 5,882,869 |
| Length | 93 |
| Max. P | 0.825900 |

| Location | 5,882,776 – 5,882,869 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -16.24 |
| Energy contribution | -16.72 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5882776 93 + 27905053 UACCC-----------AGAGCAAC---UGUCGUUGCUCCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAAAAUCCGGCACGACAAAAAAUA .....-----------.(((((((---....)))))))...........((((((((((((........)))))...((...((.....)).)))))))))...... ( -24.70) >DroPse_CAF1 9385 87 + 1 UACCA-----------AGG--AAC---UGUCGCCGCUUCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAGAAUCCUGCACGACACAA---- .....-----------...--...---(((((..(((((((.......((((((.((((((........)))))).)))))).)))))....)).)))))...---- ( -21.40) >DroSim_CAF1 6525 91 + 1 UACCC-----------AGA--AAC---UGUCGUUGCUCCGCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAAAAUCCGGCACGACAAAAAAUA .....-----------...--...---((((((.(((...........((((((.((((((........)))))).))))))((.....)))))))))))....... ( -23.40) >DroEre_CAF1 6551 91 + 1 UACCC-----------AGG--AAC---UGUUGUUGCCCCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAAAAUCCGGCACGACAAAAAAUA .....-----------...--...---((((((.(((...........((((((.((((((........)))))).))))))((.....)))))))))))....... ( -23.70) >DroAna_CAF1 9160 102 + 1 GAGCCAGUCUCAAAAAAAA--AACGCUCGUCGCCGCUUCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGGAAAUCCGGUAGGA---AAAAUA ((((...............--...))))...((((.((((((.......(((((.((((((........)))))).)))))))))))...))))....---...... ( -21.58) >DroPer_CAF1 9621 87 + 1 UACCG-----------AAG--AAC---UGUCGCCGCUUCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAGAAUCCUGCACGACACAA---- .....-----------...--...---(((((..(((((((.......((((((.((((((........)))))).)))))).)))))....)).)))))...---- ( -21.40) >consensus UACCC___________AGA__AAC___UGUCGCCGCUCCUCACAUGUAAUUGUUGUGACAUUAUUUAAUAUGUCAAAGCAAUGGAAAAUCCGGCACGACAAAAAAUA ...........................(((((..(((...........((((((.((((((........)))))).))))))((.....))))).)))))....... (-16.24 = -16.72 + 0.47)

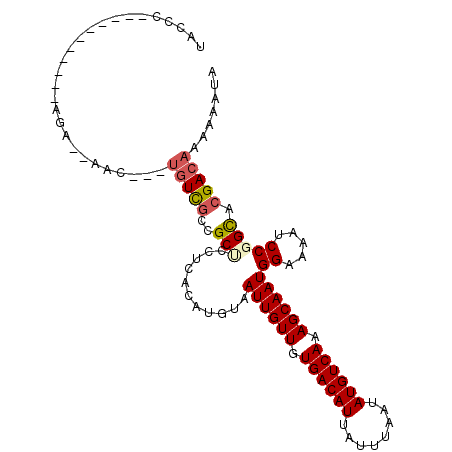

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:30 2006