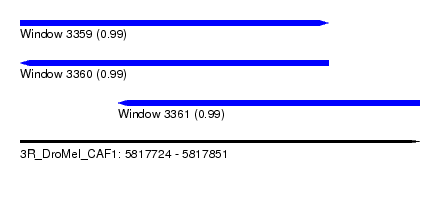
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,817,724 – 5,817,851 |
| Length | 127 |
| Max. P | 0.994785 |

| Location | 5,817,724 – 5,817,822 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -21.84 |
| Energy contribution | -24.53 |
| Covariance contribution | 2.69 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.67 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
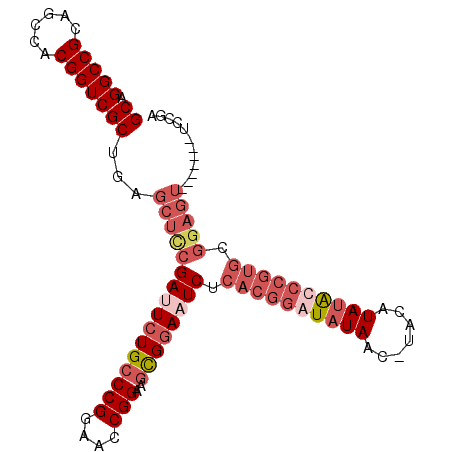
>3R_DroMel_CAF1 5817724 98 + 27905053 UUUGAGCGAAUCGGACCGUGGACUUUGGACUUUGGA-------ACUCCGCACGGGCAUAUCUAAGUUAUAUCCGUGAGAGUUCGCUUCCGGUUCCGGGCAGAAUC ((((.(.((((((((..(((((((((.......((.-------...)).(((((..(((.......)))..)))))))))))))).)))))))))...))))... ( -34.50) >DroSim_CAF1 110029 78 + 1 UUUGAGCGAAUCGGACCCUGGACUUUG---------------------------GCAUAUCUAGGUUAUAUCCGUGAGAGUCCGCUUCCGGUUCCGGGCAGAUUC ((((.(.((((((((...((((((((.---------------------------((.......((......)))).))))))))..)))))))))...))))... ( -24.71) >DroEre_CAF1 106284 94 + 1 UUUGAGCGAUUCGGACCUGGGACCUUGGACUUCGGA-------ACUCCGCACGGAUAUAUG----UUAUAUCCGUGGGAUUCCACUUCCGAUUCCGGCCAGAAUC .......(((((((.((.((((..(((((....(((-------(.(((.((((((((((..----.)))))))))))))))))...))))))))))))).))))) ( -37.90) >DroYak_CAF1 95386 105 + 1 UUUGAGCGAUUCGCACUUCGGACUUUGGACUUCGGACUUUGGUACUCCGCACGGAUGUAUGUUUGUUAUAUCCGUGAGAUUCCACUUCCGUUUCCGGGCAGAAUC .......(((((((.(....)...(((((...((((...(((...((..((((((((((.......)))))))))).))..)))..))))..))))))).))))) ( -33.50) >consensus UUUGAGCGAAUCGGACCUUGGACUUUGGACUUCGGA_______ACUCCGCACGGACAUAUCUA_GUUAUAUCCGUGAGAGUCCACUUCCGGUUCCGGGCAGAAUC ((((.(.((((((((....(((((((......((((.........))))((((((((((.......)))))))))))))))))...)))))))))...))))... (-21.84 = -24.53 + 2.69)


| Location | 5,817,724 – 5,817,822 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 73.23 |
| Mean single sequence MFE | -31.33 |
| Consensus MFE | -16.52 |
| Energy contribution | -19.02 |
| Covariance contribution | 2.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.994785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5817724 98 - 27905053 GAUUCUGCCCGGAACCGGAAGCGAACUCUCACGGAUAUAACUUAGAUAUGCCCGUGCGGAGU-------UCCAAAGUCCAAAGUCCACGGUCCGAUUCGCUCAAA ((.((.(((.(((...(((...((((((((((((.((((.......)))).))))).)))))-------)).....)))....)))..)))..)).))....... ( -28.60) >DroSim_CAF1 110029 78 - 1 GAAUCUGCCCGGAACCGGAAGCGGACUCUCACGGAUAUAACCUAGAUAUGC---------------------------CAAAGUCCAGGGUCCGAUUCGCUCAAA (((((.(((((....)))..))(((((((...((((...............---------------------------....))))))))))))))))....... ( -23.01) >DroEre_CAF1 106284 94 - 1 GAUUCUGGCCGGAAUCGGAAGUGGAAUCCCACGGAUAUAA----CAUAUAUCCGUGCGGAGU-------UCCGAAGUCCAAGGUCCCAGGUCCGAAUCGCUCAAA (((((.((((((.((((((..((((((((((((((((((.----..)))))))))).))).)-------))))...)))..))).)).)))).)))))....... ( -42.00) >DroYak_CAF1 95386 105 - 1 GAUUCUGCCCGGAAACGGAAGUGGAAUCUCACGGAUAUAACAAACAUACAUCCGUGCGGAGUACCAAAGUCCGAAGUCCAAAGUCCGAAGUGCGAAUCGCUCAAA (((((.((.((....)(((..((((....(((((((.((.......)).)))))))((((.(.....).))))...))))...)))...).)))))))....... ( -31.70) >consensus GAUUCUGCCCGGAACCGGAAGCGGAAUCUCACGGAUAUAAC_UACAUAUACCCGUGCGGAGU_______UCCGAAGUCCAAAGUCCAAGGUCCGAAUCGCUCAAA (((((...(((....)))....(((((..((((((((((.......)))))))))).(((.........))).........))))).......)))))....... (-16.52 = -19.02 + 2.50)

| Location | 5,817,755 – 5,817,851 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.39 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -27.00 |
| Energy contribution | -30.62 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.75 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988661 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5817755 96 - 27905053 GCAGGCCGCAGCCACGGUCGCUGAGCUUCGAUUCUGCCCGGAACCGGAAGCGAACUCUCACGGAUAUAACUUAGAUAUGCCCGUGCGGAGU-------UCCAA ((.(((((......)))))))...((((((.((((....)))).).)))))((((((((((((.((((.......)))).))))).)))))-------))... ( -35.40) >DroSim_CAF1 110056 80 - 1 GCAGGCCGCAGCCACGGUCGCUGAGCUUCGAAUCUGCCCGGAACCGGAAGCGGACUCUCACGGAUAUAACCUAGAUAUGC----------------------- ((.(((((......)))))))...((.((((.((((((((....)))..))))).))....((......))..))...))----------------------- ( -22.10) >DroEre_CAF1 106315 92 - 1 GCAGGCCGCAGCCACGGUCGCAGAGCUCCGAUUCUGGCCGGAAUCGGAAGUGGAAUCCCACGGAUAUAA----CAUAUAUCCGUGCGGAGU-------UCCGA ((.(((((......)))))))...(((((((((((....)))))))).)))(((((((((((((((((.----..)))))))))).))).)-------))).. ( -44.80) >DroYak_CAF1 95417 103 - 1 GCAGGCCGCAGCCACGGUCGCUGAGCUCCGAUUCUGCCCGGAAACGGAAGUGGAAUCUCACGGAUAUAACAAACAUACAUCCGUGCGGAGUACCAAAGUCCGA ...(((....))).(((.(..((.((((((((((..((((....)))..)..))))).(((((((.((.......)).))))))).)))))..))..).))). ( -41.30) >consensus GCAGGCCGCAGCCACGGUCGCUGAGCUCCGAUUCUGCCCGGAACCGGAAGCGGAAUCUCACGGAUAUAAC_UACAUAUACCCGUGCGGAGU_______UCCGA ((.(((((......)))))))...((((((((((((((((....)))..)))))))).((((((((((.......)))))))))).)))))............ (-27.00 = -30.62 + 3.62)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:26:13 2006