
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 868,732 – 868,966 |
| Length | 234 |
| Max. P | 0.844543 |

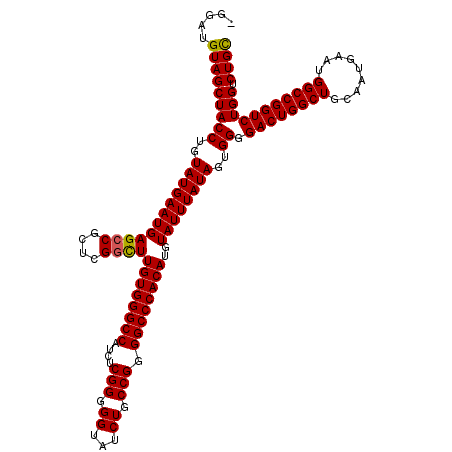
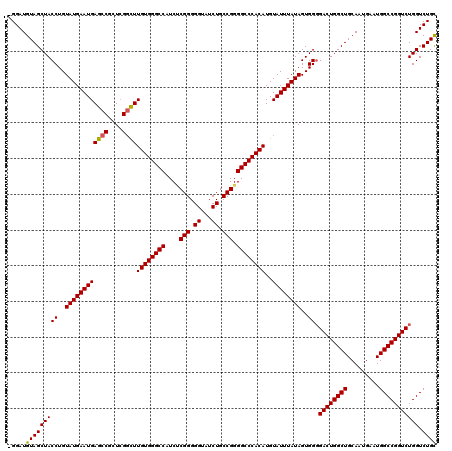
| Location | 868,732 – 868,851 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -49.33 |
| Consensus MFE | -46.40 |
| Energy contribution | -46.27 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 868732 119 - 27905053 -GGAUGUAGCUACCUGUAUGAAUGAGCCGCUCGGCUUGUGGGCCAUCUCGGGGGGAUCUGCCGGGGCCCACAUGUAUUUAUAGUGGGGACUGGCUGCAAUGAAUGGCCGGUCUGGUCUGU -((((.......((..((((((((((((....))))((((((((...((((.((...)).))))))))))))..))))))))..))(((((((((.........))))))))).)))).. ( -49.30) >DroSec_CAF1 43750 118 - 1 -GGAUGUAGCUACCUGUAUGAAUGAGCCGCUGGGCUUGUGGGCCAUCUCGGGGG-AUCUGCCGGGGCCCACAUGUAUUUAUAGUGGGGACUGGCUGCAAUGAAUGGCCGGUCUGGUCUGC -....(((((((((((((((((((((((....))))((((((((...((((.(.-...).))))))))))))..)))))))).))))((((((((.........))))))))))).)))) ( -49.10) >DroSim_CAF1 42962 119 - 1 -GGAUGUAGCUACCUGUAUGAAUGAGCCGCUCGGCUUGUGGGCCAUCCCGGGGGUAUCUGCCGGGGCCCACAUGUAUUUAUAGUGGGGACUGGCUGCAAUGAAUGGCCGGUCUGGUCUGC -....(((((((((((((((((((((((....))))((((((((...((((.((...)).))))))))))))..)))))))).))))((((((((.........))))))))))).)))) ( -52.70) >DroEre_CAF1 42986 119 - 1 UGGAUGUAGCUACCUGUAUGAAUGAGCCGCUCGCUUUGUGGGCCAUCACGGGGGUAUCUGCCGGGGCCCACAUGUAUUUAUAGUGG-GACUGGCUGCAAUGAAUGGCCGGUCUGGUCUGC .....(((((((((..(((((((((((.....))).((((((((....(((.((...)).))).))))))))..))))))))..))-((((((((.........))))))))))).)))) ( -46.20) >consensus _GGAUGUAGCUACCUGUAUGAAUGAGCCGCUCGGCUUGUGGGCCAUCUCGGGGGUAUCUGCCGGGGCCCACAUGUAUUUAUAGUGGGGACUGGCUGCAAUGAAUGGCCGGUCUGGUCUGC .....(((((((((..((((((((((((....))))((((((((....(((.((...)).))).))))))))..))))))))..)).((((((((.........))))))))))).)))) (-46.40 = -46.27 + -0.13)

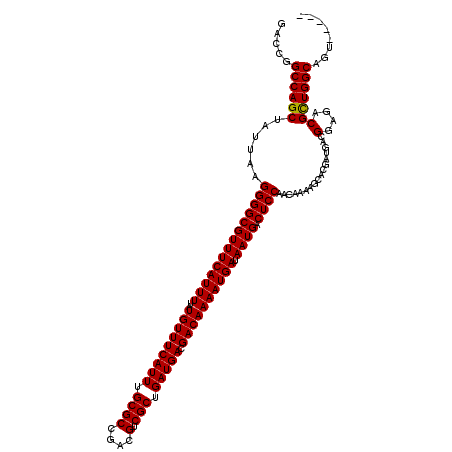
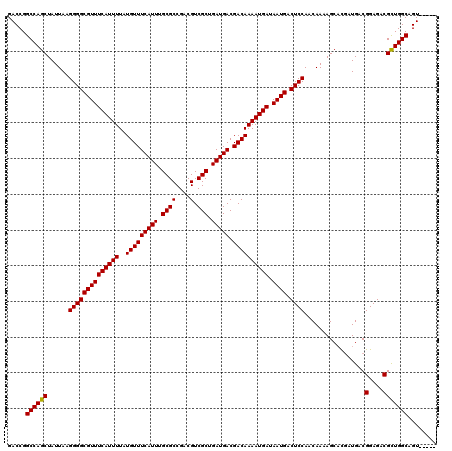
| Location | 868,851 – 868,966 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -37.22 |
| Consensus MFE | -35.99 |
| Energy contribution | -35.80 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 868851 115 - 27905053 GACCGGCCAGCUAUUAAGGGGCGUUUCAUUUUAUGUUUCAUUUGCGCCGACGUCGCUGAUGACGACAAAAUGAUAAUGACUCCAACAAAAGCACGAUGACGGAGACGCUGGCAGU----- .....((((((......((((((((((((((..(((((((((.((((....).))).))))).)))))))))).)))).)))).................(....)))))))...----- ( -36.40) >DroSec_CAF1 43868 115 - 1 GACCGGCCAGCUAUUAAGGGGCGUUUCAUUUUAUGUUUCAUUUGCGCCGACGUCGCUGAUGACGACAAAAUGAUAAUGACUCCAACAAAAGCACGAUGACGGAGACGCUGGCAGU----- .....((((((......((((((((((((((..(((((((((.((((....).))).))))).)))))))))).)))).)))).................(....)))))))...----- ( -36.40) >DroSim_CAF1 43081 115 - 1 GACCGGCCAGCUAUUAAGGGGCGUUUCAUUUUAUGUUUCAUUUGCGCCGACGUCGCUGAUGACGACAAAAUGAUAAUGACUCCAACAAAAGCACGAUGACGGAGACGCUGGCAGU----- .....((((((......((((((((((((((..(((((((((.((((....).))).))))).)))))))))).)))).)))).................(....)))))))...----- ( -36.40) >DroEre_CAF1 43105 120 - 1 GACCGGCCAGCUAUUAAGGGGCGUUUCAUUUUAUGUUUCAUUUGCGCCGACGUCGCUGAUGACGACAAAAUGAUAAUGACUCCAACAAAAGCCUCAUGACGGAGACGUUGGCAGUGGCUA ....(((((.((.....((((((((((((((..(((((((((.((((....).))).))))).)))))))))).)))).)))).......(((....((((....)))))))))))))). ( -39.70) >consensus GACCGGCCAGCUAUUAAGGGGCGUUUCAUUUUAUGUUUCAUUUGCGCCGACGUCGCUGAUGACGACAAAAUGAUAAUGACUCCAACAAAAGCACGAUGACGGAGACGCUGGCAGU_____ .....((((((......((((((((((((((..(((((((((.((((....).))).))))).)))))))))).)))).)))).................(....)))))))........ (-35.99 = -35.80 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:24 2006