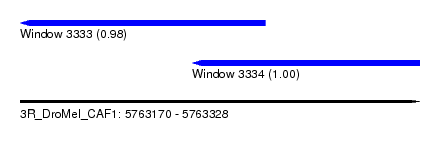
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,763,170 – 5,763,328 |
| Length | 158 |
| Max. P | 0.996798 |

| Location | 5,763,170 – 5,763,267 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 68.48 |
| Mean single sequence MFE | -26.11 |
| Consensus MFE | -7.14 |
| Energy contribution | -7.47 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
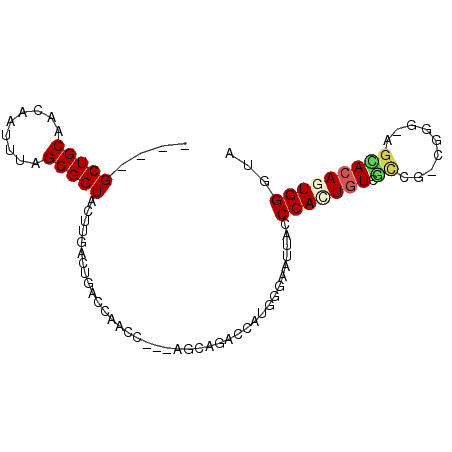
>3R_DroMel_CAF1 5763170 97 - 27905053 CCACUGUGUUCG-UGAG-AUCACAGUGGGUAAAUGGCAGCUACAAAUCUGACAUUA-----AAGGAACUUGGCAAUAACAUUUAUAUACAAAAUCUGGCUUUAA ((((((((.((.-...)-).))))))))(((((((...((((....(((.......-----.)))....)))).....)))))))................... ( -26.70) >DroPse_CAF1 60955 82 - 1 CCAGUGUCGCCUACGGGUGGCACAGUGGCAACAC--------ACACUCACACACAA-----GAGGAAGUGG-CA--------UACAUGGAAGAGAAUAUUUUAG (((((((((((....)))))))).(((....)))--------.((((..(......-----..)..)))).-..--------....)))............... ( -28.10) >DroSec_CAF1 55397 97 - 1 CCACUGUCUUCG-UGAG-AGAACAGUGUGUAAAUGGCAGCCACAAAGCUGACAUUA-----AAGGAACUAGGCAAUAAAAUGUAUAUAAAAAAUAUGGCUUUAA .((((((..((.-...)-)..))))))....((((.((((......)))).)))).-----........((((.(((................))).))))... ( -20.59) >DroSim_CAF1 55590 97 - 1 CCACUGUCUUCG-UGAG-AGAACAGUGGGUAAAUGGCAGCUACAAAGCUGACAUUA-----AAGGAACUAGGCAAUAAAAUGUAUAUAAAAAAUAUGGCUUUAA (((((((..((.-...)-)..)))))))...((((.(((((....))))).)))).-----........((((.(((................))).))))... ( -25.69) >DroEre_CAF1 50591 102 - 1 CCACUGUGUUCA-UGUG-AGAACAGUGGGUAAAUGGCUGAUACAAAUGAGGCAUUAUGGUUAGGAAACUUGGCAAGAACACUUAUAUUAAGUAGCUGGCAUUAA (((((((.(((.-...)-)).)))))))((....(((((......(((((...((.((.(.((....)).).)).))...)))))......))))).))..... ( -25.80) >DroYak_CAF1 39289 102 - 1 CCACUGUGUUCA-UGUG-AGAACAGUGGGCAAAUGGCUGCCACAAAUGAGGCAUUAUGGCUAAGAGACUUGGCAAGAACAAUCAUAUAAAGUAUGUGGCAUUAA (((((((.(((.-...)-)).)))))))((.....))(((((((......((.(((((((((((...))))))..((....)).))))).)).))))))).... ( -29.80) >consensus CCACUGUCUUCG_UGAG_AGAACAGUGGGUAAAUGGCAGCUACAAAUCAGACAUUA_____AAGGAACUUGGCAAUAACAUUUAUAUAAAAAAUAUGGCUUUAA (((((((..............)))))))............................................................................ ( -7.14 = -7.47 + 0.33)



| Location | 5,763,238 – 5,763,328 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 73.11 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.35 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.33 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5763238 90 - 27905053 UCCUGCUGCAACAAUUUAGCGGUACUUGACUGACCAGCCGGCAGCAGACCAUGGGAAUUGCCCACUGUGUUCG-UGAG-AUCACAGUGGGUA ((((((((........))))(((...((.(((.((....))))))).)))..))))..(((((((((((.((.-...)-).))))))))))) ( -39.50) >DroPse_CAF1 61006 79 - 1 ----GCUGCAACGAUUUAGCGGUACUUG------CAACC---GCAAGACCAUGGGAAUUACCCAGUGUCGCCUACGGGUGGCACAGUGGCAA ----((..(...........(((.((((------(....---)))))))).((((.....))))((((((((....)))))))).)..)).. ( -37.80) >DroSim_CAF1 55658 90 - 1 UGCUGCUGCAACAAUUUAGCGGUACUUGACUGACCAGCCGGCAGCAGACCAUGGGAAUUGCCCACUGUCUUCG-UGAG-AGAACAGUGGGUA ..(((((((..........((((.....))))........)))))))...........((((((((((..((.-...)-)..)))))))))) ( -36.87) >DroEre_CAF1 50664 87 - 1 ---UGCUGCAACAAUUUAGCGGUACUUGACUGGCCGACCAGCAGCAGACCAUGGGAAUUGCCCACUGUGUUCA-UGUG-AGAACAGUGGGUA ---.((((........))))(((..(((.((((....)))))))...)))........((((((((((.(((.-...)-)).)))))))))) ( -31.00) >DroAna_CAF1 6701 75 - 1 ----GCUGC-ACAAUUUAGCGGUACUCGGCUGACCA----------GGCCAUGGGAAAUACCCAUUAUAGCCGCCGGG-AGUACAGUGGUU- ----((((.-((..((..(((((....((((.....----------))))(((((.....)))))....)))))..))-.)).))))....- ( -24.90) >DroPer_CAF1 60483 79 - 1 ----GCUGCAACGAUUUAGCGGUACUUG------CAACC---ACAAGACCAUGGGAAUUACCCAGUGUCGCCUACGGGUGGCACAGUGGCAA ----((..(...........(((.((((------.....---.))))))).((((.....))))((((((((....)))))))).)..)).. ( -34.20) >consensus ____GCUGCAACAAUUUAGCGGUACUUGACUGACCAACC___AGCAGACCAUGGGAAUUACCCACUGUCGCCG_CGGG_AGCACAGUGGGUA ....(((((.........)))))......................................(((((((.((.........)))))))))... (-11.15 = -11.35 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:46 2006