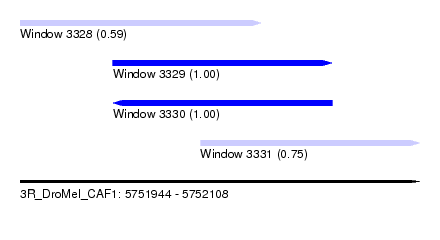
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,751,944 – 5,752,108 |
| Length | 164 |
| Max. P | 0.999941 |

| Location | 5,751,944 – 5,752,043 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.55 |
| Mean single sequence MFE | -31.31 |
| Consensus MFE | -15.73 |
| Energy contribution | -16.51 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5751944 99 + 27905053 AUUUGAAAGUCUUUGACUUGCUGCAGUUAGCUCAAACAAAAUAAAAACUGCCU----CGGCCCAAGGUAAUGGGCAAUUAGCUGCAG---------------CAACCUUCUGGGCCAA ........((((..((.((((((((((((((..................))..----..(((((......)))))...)))))))))---------------)))...)).))))... ( -29.87) >DroPse_CAF1 49179 110 + 1 AUUUGAAAGUCUUUGAGUU---GCAGUUAGCUCGA----AAUAA-AAUUGCCGGUACCGAGGCAGGGUAAUGGGCAAUUAGCUGCAGCAGCAGCAACAACAGCAACCUUUUGGGCCUG ........((((..(((((---((.(((.......----.....-(((((((.(((((.......))).)).))))))).(((((....)))))...))).)))).)))..))))... ( -34.20) >DroSim_CAF1 44319 99 + 1 AUUUGAAAGUCUUUGACUUGCUGCAGUUAGCUCAAACAAAAUAAAAACUGCCU----CGGCCCAAGGUAAUGGGCAAUUAGCUGCAG---------------CAACCUUCUGGGCCAA ........((((..((.((((((((((((((..................))..----..(((((......)))))...)))))))))---------------)))...)).))))... ( -29.87) >DroEre_CAF1 39188 99 + 1 AUUUGAAAGUCUUUGACUUGCUGCAGUUAGCUCAAACAAAAUAAAAACUGCCU----CGGCCCAAGGUAAUGGGCAAUUAGCUGCAG---------------CAACCUUCUGGGCCAA ........((((..((.((((((((((((((..................))..----..(((((......)))))...)))))))))---------------)))...)).))))... ( -29.87) >DroYak_CAF1 27654 99 + 1 AUUUGAAAGUCUUUGACUUGCUGCAGUUAGCUCAAACAAAAUAAAAACUGCCU----CGGCCCAAGGUAAUGGGCAAUUAGCUGCAG---------------CAACCUUCUGGGCCAA ........((((..((.((((((((((((((..................))..----..(((((......)))))...)))))))))---------------)))...)).))))... ( -29.87) >DroPer_CAF1 48826 110 + 1 AUUUGAAAGUCUUUGAGUU---GCAGUUAGCUCGA----AAUAA-AAUUGCCGGUACCGAGGCAGGGUAAUGGGCAAUUAGCUGCAGCAGCAGCAACAACAGCAACCUUUUGGGCCUG ........((((..(((((---((.(((.......----.....-(((((((.(((((.......))).)).))))))).(((((....)))))...))).)))).)))..))))... ( -34.20) >consensus AUUUGAAAGUCUUUGACUUGCUGCAGUUAGCUCAAACAAAAUAAAAACUGCCU____CGGCCCAAGGUAAUGGGCAAUUAGCUGCAG_______________CAACCUUCUGGGCCAA ......(((((...))))).((((((((((((((..............((((.............)))).)))))...)))))))))..................((....))..... (-15.73 = -16.51 + 0.78)



| Location | 5,751,982 – 5,752,072 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -23.53 |
| Energy contribution | -23.53 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5751982 90 + 27905053 AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCA----------ACAUGGGCUAGUCAAA ................((((((((((..(((((.....))).))...))))..))))))..........((((.----------.....))))....... ( -25.10) >DroSec_CAF1 43243 90 + 1 AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCA----------ACAUGGGCUAGUCAAA ................((((((((((..(((((.....))).))...))))..))))))..........((((.----------.....))))....... ( -25.10) >DroSim_CAF1 44357 90 + 1 AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCA----------ACAUGGGCUAGUCAAA ................((((((((((..(((((.....))).))...))))..))))))..........((((.----------.....))))....... ( -25.10) >DroEre_CAF1 39226 90 + 1 AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCA----------ACAUGGGCUAGUCAAA ................((((((((((..(((((.....))).))...))))..))))))..........((((.----------.....))))....... ( -25.10) >DroYak_CAF1 27692 100 + 1 AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCAACAGCCAACAACAUGGGCUAGUCAAA ........(((((((.((((((((((..(((((.....))).))...))))..))))))...........((.....)).........)))).))).... ( -26.10) >consensus AAAUAAAAACUGCCUCGGCCCAAGGUAAUGGGCAAUUAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCA__________ACAUGGGCUAGUCAAA ........((((((..((((((((((..(((((.....))).))...))))..)))))).........((.................))))).))).... (-23.53 = -23.53 + -0.00)


| Location | 5,751,982 – 5,752,072 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -27.19 |
| Energy contribution | -27.19 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5751982 90 - 27905053 UUUGACUAGCCCAUGU----------UGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(..((.((((.....----------.))))))..).((((((((((..((((....((((....)))).....))))))))))))))............ ( -28.80) >DroSec_CAF1 43243 90 - 1 UUUGACUAGCCCAUGU----------UGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(..((.((((.....----------.))))))..).((((((((((..((((....((((....)))).....))))))))))))))............ ( -28.80) >DroSim_CAF1 44357 90 - 1 UUUGACUAGCCCAUGU----------UGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(..((.((((.....----------.))))))..).((((((((((..((((....((((....)))).....))))))))))))))............ ( -28.80) >DroEre_CAF1 39226 90 - 1 UUUGACUAGCCCAUGU----------UGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(..((.((((.....----------.))))))..).((((((((((..((((....((((....)))).....))))))))))))))............ ( -28.80) >DroYak_CAF1 27692 100 - 1 UUUGACUAGCCCAUGUUGUUGGCUGUUGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(((.((.(((((.((.((.(((.(((((((((.(((....((((.....)))))))))))))))).))).)).))..)))))..)).)))......... ( -31.80) >consensus UUUGACUAGCCCAUGU__________UGGCUGUUGGUUUUUGGCCCAGAAGGUUGCUGCAGCUAAUUGCCCAUUACCUUGGGCCGAGGCAGUUUUUAUUU .(..((.((((................))))))..).((((((((((..((((....((((....)))).....))))))))))))))............ (-27.19 = -27.19 + -0.00)



| Location | 5,752,018 – 5,752,108 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 90 |
| Reading direction | forward |
| Mean pairwise identity | 91.85 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -20.85 |
| Energy contribution | -20.63 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5752018 90 + 27905053 UAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCAACAUGGGCUAGUCAAACAAAUGAGGCAAGCGGACAGCAAACUCCGGACCUGG ..(((((.((..((....))(((..........((((......))))..(((......))))))..)).).)))).....(((....))) ( -22.10) >DroSec_CAF1 43279 90 + 1 UAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCAACAUGGGCUAGUCAAACAAAUGAGGCAAGCGUACAGUAAACUCCGGACCAGC ..((((..((..((....))(((..........((((......))))..(((......))))))..))...))))............... ( -19.10) >DroEre_CAF1 39262 82 + 1 UAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCAACAUGGGCUAGUCAAACAAAUGAGGCAAGCGGACAGCAAACUCC-------- ..(((((.((..((....))(((..........((((......))))..(((......))))))..)).).)))).......-------- ( -21.80) >consensus UAGCUGCAGCAACCUUCUGGGCCAAAAACCAACAGCCAACAUGGGCUAGUCAAACAAAUGAGGCAAGCGGACAGCAAACUCCGGACC_G_ ..((((..((..((....))(((..........((((......))))..(((......))))))..))...))))............... (-20.85 = -20.63 + -0.22)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:43 2006