
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,750,187 – 5,750,322 |
| Length | 135 |
| Max. P | 0.993000 |

| Location | 5,750,187 – 5,750,284 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.64 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -22.33 |
| Energy contribution | -22.41 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
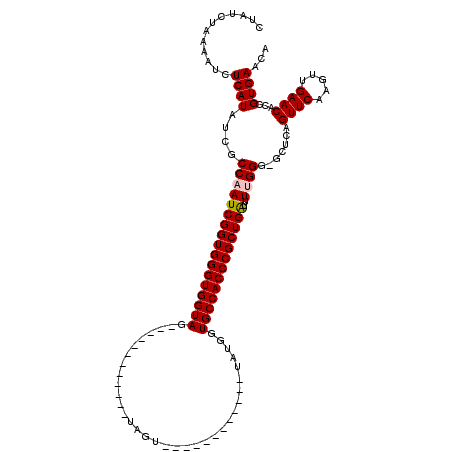
>3R_DroMel_CAF1 5750187 97 + 27905053 UUUGGUCAUGUGUCAAUUCCCGCACCUCUUGUUUGCAUAAUGUUCACAGUGUUCAACUUGAACUGAGC-CCCAAAAUCAGCGGGUGGCACCAUA------------ACUA---------- ..((((.((((((((...(((((.....(((...((.....(((((.(((.....))))))))...))-..))).....)))))))))).))).------------))))---------- ( -24.90) >DroSec_CAF1 41393 98 + 1 UUUGGUCAUGUGUCAAUUCCCGCGCCUCUUGUUUGCAUAAUGUUCACGGUGUUCAACUUGAACUGAGCACCCAAAGCCAGCGGGUGGCACCAUA------------ACUA---------- ..((((.((((((((...(((((....(((((..((.....))..))((((((((........))))))))..)))...)))))))))).))).------------))))---------- ( -31.00) >DroSim_CAF1 42529 98 + 1 UUUGGUCAUGUGUCAAUUCCCGCGCCUCUUGUUUGCAUAAUGUUCACGGUGUUCAACUUGAACUGAGCACCCAAAACCAGCGGGUGGCACCAUA------------ACUA---------- ..((((.((((((((...(((((.......((..((.....))..))((((((((........))))))))........)))))))))).))).------------))))---------- ( -31.00) >DroEre_CAF1 37420 118 + 1 UUUGGUCAUGUGUCAAUUCCCGCGCCUCUUGUUUGCAUAAUGUUCACGGUGUUCAACUUGAACUGCGC--CCCAAGUCAGCGGGUGGCACUGUAACUACCACUAUAACUACCACCUUAAC ..((((((.((((((...(((((....((((..((((....(((((.(((.....)))))))))))).--..))))...))))))))))))).....))))................... ( -31.30) >DroYak_CAF1 25742 108 + 1 UUUGGUCAUGUGUCAAUUCCCGCGCCUCUUGUUUGCAUAAUGUUCACCGUGUUCAACUUGAACUGAGC--CCCAAAUCAGCGGGUGGCACUGUAACUACCACUAUAACUA---------- ..((((((.((((((...(((((((.........)).....(((((....((((.....)))))))))--.........))))))))))))).....)))).........---------- ( -25.90) >consensus UUUGGUCAUGUGUCAAUUCCCGCGCCUCUUGUUUGCAUAAUGUUCACGGUGUUCAACUUGAACUGAGC_CCCAAAAUCAGCGGGUGGCACCAUA____________ACUA__________ .........((((((...(((((...........((.....(((((.(((.....))))))))...))...........))))))))))).............................. (-22.33 = -22.41 + 0.08)



| Location | 5,750,227 – 5,750,322 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.18 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.60 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5750227 95 - 27905053 CUAUCUAAAAUCUCAUAUCGCCAAUCGGUGGGUGGUAG------------UAGU------------UAUGGUGCCACCCGCUGAUUUUGGG-GCUCAGUUCAAGUUGAACACUGUGAACA ............(((((..((((((((((((((((((.------------((..------------..)).)))))))))))))))....)-))...((((.....))))..)))))... ( -37.00) >DroSec_CAF1 41433 96 - 1 CUAUCUAAAAUCUCAUAUCGCCAAUCGGUGGGUGGUAG------------UAGU------------UAUGGUGCCACCCGCUGGCUUUGGGUGCUCAGUUCAAGUUGAACACCGUGAACA .................((((((((((((((((((((.------------((..------------..)).)))))))))))))..)))((((.((((......)))).))))))))... ( -36.00) >DroSim_CAF1 42569 96 - 1 CUAUCUAAAAUCUCAUAUCGCCAAUCGGUGGGUGGUAG------------UAGU------------UAUGGUGCCACCCGCUGGUUUUGGGUGCUCAGUUCAAGUUGAACACCGUGAACA .................((((.(((((((((((((((.------------((..------------..)).)))))))))))))))...((((.((((......)))).))))))))... ( -37.40) >DroEre_CAF1 37460 118 - 1 CUAUCUGCAAUCUCAUAUCGCCAAUCGGUGGGUGGUAGCAGUUAAGGUGGUAGUUAUAGUGGUAGUUACAGUGCCACCCGCUGACUUGGG--GCGCAGUUCAAGUUGAACACCGUGAACA ............((((..((((..(((((((((((((((.......)).(((..(((....)))..)))..))))))))))))).....)--)))..((((.....))))...))))... ( -37.00) >DroYak_CAF1 25782 106 - 1 CUAUCUGAAAUCUCAUAUCGCCAAUCGGUGGGUGGUAG------------UAGUUAUAGUGGUAGUUACAGUGCCACCCGCUGAUUUGGG--GCUCAGUUCAAGUUGAACACGGUGAACA .................(((((((((((((((((((((------------((..(((....)))..)))..)))))))))))))))....--.....((((.....))))..)))))... ( -38.60) >consensus CUAUCUAAAAUCUCAUAUCGCCAAUCGGUGGGUGGUAG____________UAGU____________UAUGGUGCCACCCGCUGAUUUUGGG_GCUCAGUUCAAGUUGAACACCGUGAACA ............((((....(((((((((((((((((..................................)))))))))))))..)))).......((((.....))))...))))... (-27.44 = -27.60 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:40 2006