| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,745,671 – 5,745,804 |
| Length | 133 |
| Max. P | 0.940378 |

| Location | 5,745,671 – 5,745,778 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 91.87 |
| Mean single sequence MFE | -39.56 |
| Consensus MFE | -34.72 |
| Energy contribution | -34.00 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5745671 107 - 27905053 UAUGCCGUGUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCACUGGCGACUGGCAC----CACUGGCACUUGUUGGAAAUAGCUUAAAGCCGGGCUCGUGCAAACUAAAGU ((((((((((...))))))))))..(((((((((....))).(((((....(((..----..(..((....))..).....)))....)))))..)))))).......... ( -38.30) >DroSec_CAF1 36965 107 - 1 UAUGCCGAGUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCGCUGGCGACUGGCAC----CACUGGCACUUGUUGGAAAUAGCUUAAAGCCGGGCUCGUGCAAACUAAAGU ..((((.(((.((..((((((.((.((.....)).))))))))..)).))))))).----..(((((....((((....)))).....)))))((....)).......... ( -38.70) >DroSim_CAF1 38064 107 - 1 UAUGCCGAGUGACAUGCGGCAUGAGGCAUGGGGCAACUGCCGCUGGCGACUGGCAC----CACUGGCACUUGUUGGAAAUAGCUUAAAGCCGGGCUCGUGCAAACUAAAGU ..((((((((..(((((........))))).(((....))).(((((....(((..----..(..((....))..).....)))....)))))))))).)))......... ( -37.00) >DroEre_CAF1 32838 110 - 1 UAUGCCGAGUGGCAUGCGGCAUUUGGCAUGGGGCAACUGCCACUGGCAGCUGGCACCA-CCACUGGCACUUGUUGGAAAUAACUUAAAGCCGGGCUCGUGGAAACUAAAGU (((((((((((.(....).))))))))))).(.((.(((((...))))).)).).(((-(..(((((....((((....)))).....)))))....)))).......... ( -40.70) >DroYak_CAF1 20930 111 - 1 UAUGCCGCGUGGCAUGCGGCAUUUGGCAUGGGGCAACUGCCACCGGCAACUGCCACUGGCAACUGGCACUUGUUGGAAAUAACUUAAAGCCGGGCUCGUGGAAACCAAAGU ....(((((.(((...((((...(((((...(....))))))(((((((.(((((..(....)))))).)))))))............)))).)))))))).......... ( -43.10) >consensus UAUGCCGAGUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCACUGGCGACUGGCAC____CACUGGCACUUGUUGGAAAUAGCUUAAAGCCGGGCUCGUGCAAACUAAAGU ..(((((.(((((((((........)))).((....))))))))))))....((((......(((((....((((....)))).....)))))....)))).......... (-34.72 = -34.00 + -0.72)

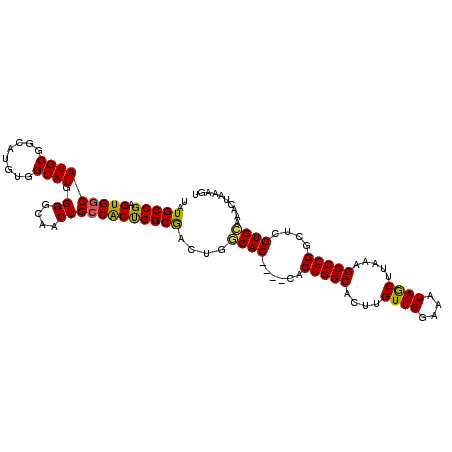

| Location | 5,745,711 – 5,745,804 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -24.27 |
| Energy contribution | -24.22 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.77 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5745711 93 + 27905053 AAGUGCCAGUG----GUGCCAGUCG-CCAGUGGCAGUUGCCCCAUGCCACAUGCCGCAUGCCAC----A----CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ...(((((.((----(((.....))-))).)))))(((((((..(((((.((((((........----.----))))))....)))))...)))))))........ ( -43.30) >DroSec_CAF1 37005 93 + 1 AAGUGCCAGUG----GUGCCAGUCG-CCAGCGGCAGUUGCCCCAUGCCACAUGCCGCAUGCCAC----U----CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ...((((..((----(((.....))-)))..))))(((((((..(((((.((((((........----.----))))))....)))))...)))))))........ ( -38.50) >DroSim_CAF1 38104 93 + 1 AAGUGCCAGUG----GUGCCAGUCG-CCAGCGGCAGUUGCCCCAUGCCUCAUGCCGCAUGUCAC----U----CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ...((((..((----(((.....))-)))..))))(((((((..((((..((((((........----.----)))))).....))))...)))))))........ ( -37.40) >DroEre_CAF1 32878 96 + 1 AAGUGCCAGUGG-UGGUGCCAGCUG-CCAGUGGCAGUUGCCCCAUGCCAAAUGCCGCAUGCCAC----U----CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ...(((((.(((-..((....))..-))).)))))(((((((..(((((.((((((........----.----))))))....)))))...)))))))........ ( -46.00) >DroYak_CAF1 20970 97 + 1 AAGUGCCAGUUGCCAGUGGCAGUUG-CCGGUGGCAGUUGCCCCAUGCCAAAUGCCGCAUGCCAC----G----CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ..........(((((.((((....)-))).)))))(((((((..(((((.(((((((.......----)----))))))....)))))...)))))))........ ( -47.70) >DroPer_CAF1 42665 96 + 1 AAGUGCCU--C----AUGCUCAUGCCCCAGCCCCAG----CCCAUGCCACAUGCCGCAUGGCCCCUACUCUACUGCCGUAUCAUAGCAAAUGGGCAGAAAUUUCAA ...(((((--.----.((((.((((..(((....((----.((((((........))))))...))......)))..))))...))))...))))).......... ( -21.20) >consensus AAGUGCCAGUG____GUGCCAGUCG_CCAGCGGCAGUUGCCCCAUGCCACAUGCCGCAUGCCAC____U____CGGCAUAUCAUGGCAAAUGGGCAACAAUUUGAA ...((((.........(((((........((((((...((.....))....))))))(((((............)))))....)))))....)))).......... (-24.27 = -24.22 + -0.05)


| Location | 5,745,711 – 5,745,804 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.27 |
| Mean single sequence MFE | -40.18 |
| Consensus MFE | -23.82 |
| Energy contribution | -25.79 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.880942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5745711 93 - 27905053 UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG----U----GUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCACUGG-CGACUGGCAC----CACUGGCACUU ........(((((((...((((((...((((((----(----((...)))))))))))))))..)))))))(((((.(((-.(.....).)----)).)))))... ( -43.80) >DroSec_CAF1 37005 93 - 1 UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG----A----GUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCGCUGG-CGACUGGCAC----CACUGGCACUU ........(((((((...((((((...((((((----.----........))))))))))))..)))))))(((((.(((-.(.....).)----)).)))))... ( -40.60) >DroSim_CAF1 38104 93 - 1 UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG----A----GUGACAUGCGGCAUGAGGCAUGGGGCAACUGCCGCUGG-CGACUGGCAC----CACUGGCACUU ........(((((((...((((....(((((((----.----........))))))).))))..)))))))(((((.(((-.(.....).)----)).)))))... ( -38.30) >DroEre_CAF1 32878 96 - 1 UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG----A----GUGGCAUGCGGCAUUUGGCAUGGGGCAACUGCCACUGG-CAGCUGGCACCA-CCACUGGCACUU ........(((((((...(((((....((((((----.----........)))))).)))))..)))))))(((((.(((-....((....))-))).)))))... ( -40.00) >DroYak_CAF1 20970 97 - 1 UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG----C----GUGGCAUGCGGCAUUUGGCAUGGGGCAACUGCCACCGG-CAACUGCCACUGGCAACUGGCACUU ........(((((((...(((((....((((((----(----((...))))))))).)))))..)))))))(((((..((-(....)))..))))).......... ( -47.20) >DroPer_CAF1 42665 96 - 1 UUGAAAUUUCUGCCCAUUUGCUAUGAUACGGCAGUAGAGUAGGGGCCAUGCGGCAUGUGGCAUGGG----CUGGGGCUGGGGCAUGAGCAU----G--AGGCACUU ......(..(.((((.....((((..(((....)))..))))((.((((((.(....).)))))).----)).)))).)..).....((..----.--..)).... ( -31.20) >consensus UUCAAAUUGUUGCCCAUUUGCCAUGAUAUGCCG____A____GUGGCAUGCGGCAUGUGGCAUGGGGCAACUGCCACUGG_CGACUGGCAC____CACUGGCACUU ........(((((((...(((((((.(((((((..........)))))))...)))..))))..)))))))(((((.((................)).)))))... (-23.82 = -25.79 + 1.97)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:36 2006