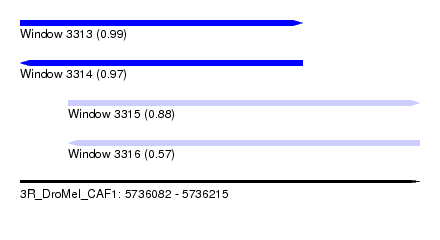
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,736,082 – 5,736,215 |
| Length | 133 |
| Max. P | 0.990386 |

| Location | 5,736,082 – 5,736,176 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -35.43 |
| Consensus MFE | -34.76 |
| Energy contribution | -34.10 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5736082 94 + 27905053 UGGCUGGCUCAUUGGCUUCAGUAUUGGGCCAAUGAUUGGCUAACCGAGCCCUGCAUGUCACUUGAUCGAUGGUUACAAAGCCAUAAGCCAUCAU ((((((((((((((((((.......))))))))))..((((.....))))......))).........((((((....)))))).))))).... ( -31.10) >DroSec_CAF1 27653 94 + 1 UGGCUGGCUCAUUGGCUCCAUUAUUGGGCCAAUGAUUGGCUAACCGAGCCCUGCAUGGCACUUGAUCGAUGGCUGCACGGCCAUAAGCCAUCAU ((((..(.((((((((.(((....))))))))))))..)))).....(((......)))........(((((((...........))))))).. ( -38.80) >DroSim_CAF1 28289 94 + 1 UGGCUGGCUCAUUGGCUCCAUUAUUGGGCCAAUGAUUGGCUAACCGAGCCCUGCAUGGCACUUGAUCGAUGGUUGCACGGCCAUAAGCCAUCAU ((((..(.((((((((.(((....))))))))))))..)))).....(((......)))........(((((((...........))))))).. ( -36.40) >consensus UGGCUGGCUCAUUGGCUCCAUUAUUGGGCCAAUGAUUGGCUAACCGAGCCCUGCAUGGCACUUGAUCGAUGGUUGCACGGCCAUAAGCCAUCAU ((((((((((((((((.(((....)))))))))))((((....)))))))..................((((((....)))))).))))).... (-34.76 = -34.10 + -0.66)



| Location | 5,736,082 – 5,736,176 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.04 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -32.47 |
| Energy contribution | -32.37 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5736082 94 - 27905053 AUGAUGGCUUAUGGCUUUGUAACCAUCGAUCAAGUGACAUGCAGGGCUCGGUUAGCCAAUCAUUGGCCCAAUACUGAAGCCAAUGAGCCAGCCA ....(((((((((((((((((............((.....)).(((((.((((....))))...)))))..))).)))))).)))))))).... ( -29.90) >DroSec_CAF1 27653 94 - 1 AUGAUGGCUUAUGGCCGUGCAGCCAUCGAUCAAGUGCCAUGCAGGGCUCGGUUAGCCAAUCAUUGGCCCAAUAAUGGAGCCAAUGAGCCAGCCA .(((((((((((....))).)))))))).....(.(((......))).)((((.((...(((((((((((....))).)))))))))).)))). ( -38.70) >DroSim_CAF1 28289 94 - 1 AUGAUGGCUUAUGGCCGUGCAACCAUCGAUCAAGUGCCAUGCAGGGCUCGGUUAGCCAAUCAUUGGCCCAAUAAUGGAGCCAAUGAGCCAGCCA ....(((((..(((((.((((..(((.......)))...)))).))).))...))))).(((((((((((....))).))))))))........ ( -36.10) >consensus AUGAUGGCUUAUGGCCGUGCAACCAUCGAUCAAGUGCCAUGCAGGGCUCGGUUAGCCAAUCAUUGGCCCAAUAAUGGAGCCAAUGAGCCAGCCA ....(((((..(((((.((((..(((.......)))...)))).))).))...))))).(((((((((((....))).))))))))........ (-32.47 = -32.37 + -0.11)



| Location | 5,736,098 – 5,736,215 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -19.67 |
| Energy contribution | -21.12 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5736098 117 + 27905053 UUCAGUAUUGGGCCAAUGAUUGGCUAACCGAGCCC--UGCAUGUCACUUGAUCGAUGGUUACAAAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUC ......(((((((((.....)))))....(((((.--.((((((....((((.(((((((...........)))))))))))(((....))).....))-)))).))))).....)))). ( -32.40) >DroSec_CAF1 27669 117 + 1 UCCAUUAUUGGGCCAAUGAUUGGCUAACCGAGCCC--UGCAUGGCACUUGAUCGAUGGCUGCACGGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUC .(((....)))((((.((...((((.....)))).--..))))))....(((((((((((....))))).((((((.(((..(((....))).......-))).))))))....)))))) ( -40.10) >DroSim_CAF1 28305 117 + 1 UCCAUUAUUGGGCCAAUGAUUGGCUAACCGAGCCC--UGCAUGGCACUUGAUCGAUGGUUGCACGGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUCACUUCGAUC .(((....)))((((.((...((((.....)))).--..))))))....(((((((((((....))))).((((((.(((..(((....))).......-))).))))))....)))))) ( -37.60) >DroEre_CAF1 23357 109 + 1 UUCAUUAUUGG-CCAA-------ACAACCGAGCCC--UGCAUGGCACUUGAUCGAUGGUUCCACAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUUACUCCGAUC .((......((-....-------....))(((((.--.(((((((...((((..(((((......))))).....))))...)))))))((((......-.)))))))))......)).. ( -27.90) >DroYak_CAF1 11118 109 + 1 UUCAGUAUUGG-CCAA-------CCAACCGAGCCC--UGCAUGGCACUUGAUCGAUGGCUCCAAAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU-AUGCUGGCUUUACUUCGAUC .((((((((((-....-------))))....(((.--.(((((((...((((..((((((....)))))).....))))...)))))))))).......-.))))))............. ( -33.30) >DroPer_CAF1 33328 101 + 1 -UCAUUAGCAGG-----------------AGGCCCUGUGCAU-ACAUUUGAUAGAUGGAUGCCGAGCACUAAGUCAUCAUUAGCUCGAUACCCAUUUAUCAUGAUCAACGGACUCUGAUU -......(((((-----------------....)))))....-.....((((((((((.((.(((((...............))))).)).)))))))))).(((((........))))) ( -27.16) >consensus UUCAUUAUUGGGCCAA_______CUAACCGAGCCC__UGCAUGGCACUUGAUCGAUGGUUGCACAGCCAUAAGCCAUCAUUAGCCAUGUGGCAAUUUAU_AUGCUGGCUUCACUUCGAUC .............................(((((....(((((((...((((.(((((((...........))))))))))))))))))((((........))))))))).......... (-19.67 = -21.12 + 1.45)


| Location | 5,736,098 – 5,736,215 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.78 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -18.04 |
| Energy contribution | -19.16 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5736098 117 - 27905053 GAUCGAAGUGAAGCCAGCAU-AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCUUUGUAACCAUCGAUCAAGUGACAUGCA--GGGCUCGGUUAGCCAAUCAUUGGCCCAAUACUGAA ......((((......(((.-......))).....((((((((((((((..((((((((((..(((.......)))...))))--)))).))...)))).))))))))))...))))... ( -34.80) >DroSec_CAF1 27669 117 - 1 GAUCGAAGUGAAGCCAGCAU-AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCCGUGCAGCCAUCGAUCAAGUGCCAUGCA--GGGCUCGGUUAGCCAAUCAUUGGCCCAAUAAUGGA .............((((((.-......))).....((((((((((((((..(((((.((((..(((.......)))...))))--.))).))...)))).))))))))))......))). ( -39.90) >DroSim_CAF1 28305 117 - 1 GAUCGAAGUGAAGCCAGCAU-AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCCGUGCAACCAUCGAUCAAGUGCCAUGCA--GGGCUCGGUUAGCCAAUCAUUGGCCCAAUAAUGGA .............((((((.-......))).....((((((((((((((..(((((.((((..(((.......)))...))))--.))).))...)))).))))))))))......))). ( -38.80) >DroEre_CAF1 23357 109 - 1 GAUCGGAGUAAAGCCAGCAU-AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCUGUGGAACCAUCGAUCAAGUGCCAUGCA--GGGCUCGGUUGU-------UUGG-CCAAUAAUGAA (((((.(((.((((((.(((-.......(((....)))..))).))))))...)))(((....))))))))..(.(((.....--.))).)((((..-------..))-))......... ( -29.80) >DroYak_CAF1 11118 109 - 1 GAUCGAAGUAAAGCCAGCAU-AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCUUUGGAGCCAUCGAUCAAGUGCCAUGCA--GGGCUCGGUUGG-------UUGG-CCAAUACUGAA ......((((..((((((..-.......(((.((((((...((((.(...((((((....))))))).))))...))))))..--.))).......)-------))))-)...))))... ( -37.69) >DroPer_CAF1 33328 101 - 1 AAUCAGAGUCCGUUGAUCAUGAUAAAUGGGUAUCGAGCUAAUGAUGACUUAGUGCUCGGCAUCCAUCUAUCAAAUGU-AUGCACAGGGCCU-----------------CCUGCUAAUGA- ...(((.((((..((..(((((((.((((((..(((((...(((....)))..)))))..)))))).)))))..)).-...))..))))..-----------------.))).......- ( -28.10) >consensus GAUCGAAGUGAAGCCAGCAU_AUAAAUUGCCACAUGGCUAAUGAUGGCUUAUGGCUGUGCAACCAUCGAUCAAGUGCCAUGCA__GGGCUCGGUUAG_______UUGGCCCAAUAAUGAA ((((((....((((((.(((........(((....)))..))).)))))).(((........)))))))))..............(((((................)))))......... (-18.04 = -19.16 + 1.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:29 2006