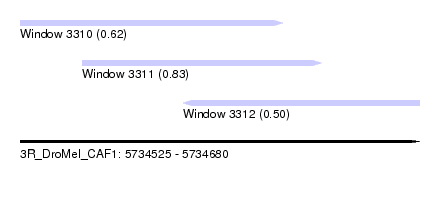
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,734,525 – 5,734,680 |
| Length | 155 |
| Max. P | 0.831196 |

| Location | 5,734,525 – 5,734,627 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.28 |
| Mean single sequence MFE | -24.14 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.622118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5734525 102 + 27905053 GCGGAAAAUGGCCAAAGAAUUUCCCUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCC ........((((((((.((.....((((....))))....((.((((((..((((....)))))))-))).))..)).))))))))...(((((....))))) ( -25.80) >DroPse_CAF1 31950 100 + 1 G-GGAAAAUGGCAAAAGAAUUUCACUCAAACGUGAGAAUAGUGGCGAAAUAAUUGAGAACGAUUUC-UGUGGCAUUUGUUUUGCCAA-AGGAAAUUGAUUUUC .-.((((((.((.......((((((......))))))......))..............(((((((-(.(((((.......))))).-.)))))))))))))) ( -23.62) >DroSim_CAF1 26706 102 + 1 GCGGAAAAUGGCCAAAGAAUUUCCCUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCC ........((((((((.((.....((((....))))....((.((((((..((((....)))))))-))).))..)).))))))))...(((((....))))) ( -25.80) >DroEre_CAF1 21719 102 + 1 GCGGAAAAUGGCCAAAGAAUUUCCCUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUAACCUGGCCAAAAGGAAAUUGAUUUCC ........((((((..........((((....))))..(((((.((....(((((....)))))..-..)).)))))...))))))...(((((....))))) ( -23.10) >DroYak_CAF1 9440 103 + 1 GCGGAAAAUGGCCAAAGAAAUUCCCUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUUGUGUGGCAUUAGCCUGGCCAAAAGGAAAUUGAUUUCC ........((((((......((((((((..(....)..))).)).)))......................(((....)))))))))...(((((....))))) ( -22.90) >DroPer_CAF1 31591 100 + 1 G-GGAAAAUGGCAAAAGAAUUUCACUCAAACGUGAGAAUAGUGGCGAAAUAAUUGAGAACGAUUUC-UGUGGCAUUUGUUUUGCCAA-AGGAAAUUGAUUUUC .-.((((((.((.......((((((......))))))......))..............(((((((-(.(((((.......))))).-.)))))))))))))) ( -23.62) >consensus GCGGAAAAUGGCCAAAGAAUUUCCCUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU_UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCC ........((((((((.((.....((((....))))....((.(((....(((((....)))))...))).))..)).))))))))...(((((....))))) (-21.23 = -21.57 + 0.34)

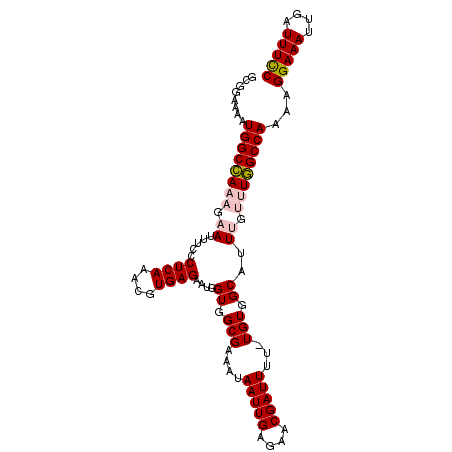
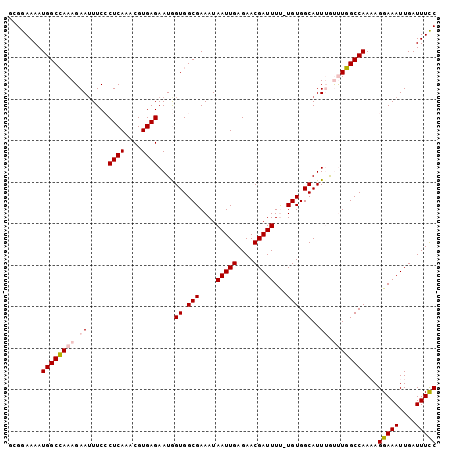
| Location | 5,734,549 – 5,734,642 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.80 |
| Mean single sequence MFE | -25.58 |
| Consensus MFE | -16.16 |
| Energy contribution | -16.80 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5734549 93 + 27905053 CUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCCAAUACUAGAA-------------------------ACAAA ((((....))))..(((((((.((((((.((...((((...)-)))..)).)))))).)))....(((((....)))))...))))...-------------------------..... ( -21.50) >DroPse_CAF1 31973 117 + 1 CUCAAACGUGAGAAUAGUGGCGAAAUAAUUGAGAACGAUUUC-UGUGGCAUUUGUUUUGCCAA-AGGAAAUUGAUUUUCAAUAAAAGAGUUUUCGUUGCCUCGGGGCCUGCCGAGGAAA ((((....))))....(..((((((..(((((((((((((((-(.(((((.......))))).-.)))))))).))))))))........))))))..)(((((......))))).... ( -38.70) >DroSec_CAF1 26120 93 + 1 CUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCCAAUACUAGAA-------------------------ACAAA ((((....))))..(((((((.((((((.((...((((...)-)))..)).)))))).)))....(((((....)))))...))))...-------------------------..... ( -21.50) >DroEre_CAF1 21743 93 + 1 CUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU-UGUGGCAUUAACCUGGCCAAAAGGAAAUUGAUUUCCUAUAGCAGAA-------------------------ACAAA ((((....))))....((.((...(((...((((.(((((((-(.((((.........))))...)))))))).)))).))).))....-------------------------))... ( -17.10) >DroYak_CAF1 9464 94 + 1 CUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUUGUGUGGCAUUAGCCUGGCCAAAAGGAAAUUGAUUUCCAAUACCAGAA-------------------------ACAAA ((((....))))..(((((((.....(((((....)))))......(((....)))..)))....(((((....)))))...))))...-------------------------..... ( -19.80) >DroPer_CAF1 31614 117 + 1 CUCAAACGUGAGAAUAGUGGCGAAAUAAUUGAGAACGAUUUC-UGUGGCAUUUGUUUUGCCAA-AGGAAAUUGAUUUUCAAUAAGAGAGUUUUCGUUGCCUAGGGGCCUGCCGAGGAAA (((...(....)......((((.....(((((((((((((((-(.(((((.......))))).-.)))))))).))))))))...............(((....))).))))))).... ( -34.90) >consensus CUCAAACGUGAGAAUGGUGGCGAAAUAAUUGAGAACGAUUUU_UGUGGCAUUUGUUUGGCCAAAAGGAAAUUGAUUUCCAAUACCAGAA_________________________ACAAA ((((....)))).....((((.((((((.((...(((......)))..)).)))))).))))...(((((....)))))........................................ (-16.16 = -16.80 + 0.64)

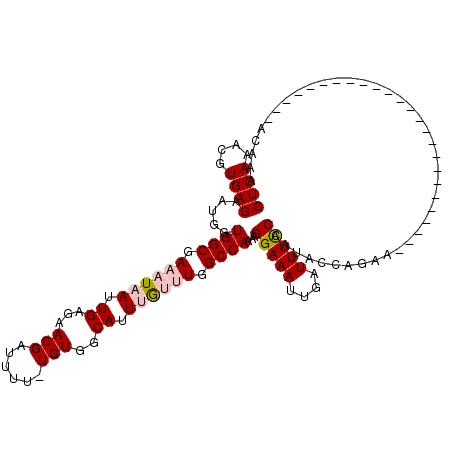
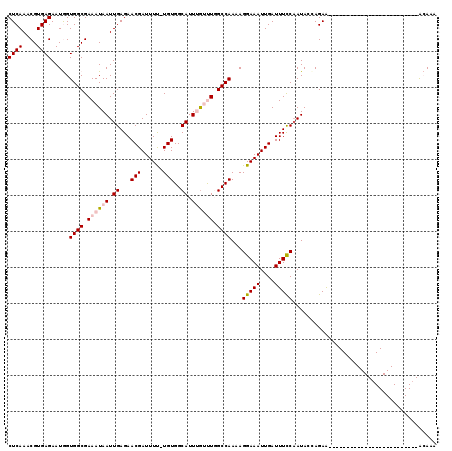
| Location | 5,734,588 – 5,734,680 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.23 |
| Mean single sequence MFE | -16.24 |
| Consensus MFE | -13.44 |
| Energy contribution | -12.24 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.83 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
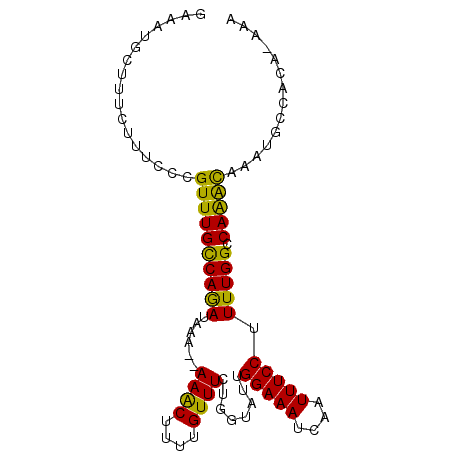
>3R_DroMel_CAF1 5734588 92 - 27905053 GAAAUGCUUUCAUUCCCGUUUGCCAGAUAAAUAAAACUUUUGUUUCUAGUAUUGGAAAUCAAUUUCCUUUUGGCCAAACAAAUGCCACA-AAA .................((((((((((......((((....))))........(((((....))))).))))).)))))..........-... ( -14.40) >DroSec_CAF1 26159 91 - 1 GAAAUGCUUUCUUUCCCGUUUGCCAAAUAAAA-AAACUUUUGUUUCUAGUAUUGGAAAUCAAUUUCCUUUUGGCCAAACAAAUGCCACA-AAA .................((((((((((.....-((((....))))........(((((....))))).))))).)))))..........-... ( -14.30) >DroSim_CAF1 26769 91 - 1 GAAAUGCUUUCUUUCCCGUUUGCCAAAUAAAC-AAGCUUUUGUUUCUGGUAUUGGAAAUCAAUUUCCUUUUGGCCAAACAAAUGCCACA-AAA .................(((((.....)))))-..((.((((((..((((...(((((....))))).....)))))))))).))....-... ( -15.90) >DroEre_CAF1 21782 92 - 1 GAAAUGCUUUCUUUCCCGUUUGCCAGAUAAAGCAAACUUUUGUUUCUGCUAUAGGAAAUCAAUUUCCUUUUGGCCAGGUUAAUGCCACA-AAA .................((((((........)))))).(((((....((((.((((((....))))))..))))..(((....))))))-)). ( -20.10) >DroYak_CAF1 9503 87 - 1 GAAAUGCUU----UCCCGUUUGUCAGAUAAA--AAACUUUUGUUUCUGGUAUUGGAAAUCAAUUUCCUUUUGGCCAGGCUAAUGCCACACAAA (((((..((----(((....(..((((..((--(....)))...))))..)..)))))...)))))..((((....(((....)))...)))) ( -16.50) >consensus GAAAUGCUUUCUUUCCCGUUUGCCAGAUAAA__AAACUUUUGUUUCUGGUAUUGGAAAUCAAUUUCCUUUUGGCCAAACAAAUGCCACA_AAA .................((((((((((......((((....))))........(((((....))))).))))).))))).............. (-13.44 = -12.24 + -1.20)

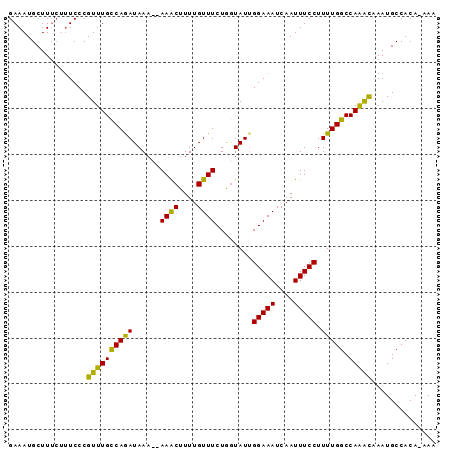
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:25 2006