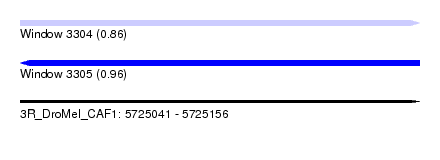
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,725,041 – 5,725,156 |
| Length | 115 |
| Max. P | 0.961947 |

| Location | 5,725,041 – 5,725,156 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -26.72 |
| Energy contribution | -28.70 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857035 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5725041 115 + 27905053 GGAGACGGCGGCAGAUUGAUGAGGCAAAUUGAUUUGCGGCUCUGCAUCUCUCAAUCUAUUGCAAAAUAGAUUGAGUGCACUAUGCCGCCGUGCUGGGUGAGAAUUGAUUGAGCCA ....(((((.((((((..((.......))..))))))(((..((((...(((((((((((....)))))))))))))))....))))))))(((.(((........))).))).. ( -44.50) >DroYak_CAF1 30207 111 + 1 GGCGACGGCGGCAGAUUGAUGAGGCAAAUUGAUUUGCGGCGCUGCAUCUCUCAAUCUAUUGCAAAA----UUGAGUGCACUCUGCCGCCGUGCUGGGUGAGAAUUGAUUGAGCCA (((.(((((((((((((((.((((((((....))))).((...))..))))))))))..((((...----.....))))....))))))))))).(((..((.....))..))). ( -41.00) >DroAna_CAF1 25535 96 + 1 AGAGAUGA-GGGAGAUUGAUGAGGCAAAUUGAUUUGCGGCGCUGCAUCUCUCAAUCUAUUGCGGAA----UUGAGUGCACU--------------GGCGAGAAUUGAUUGGGCCA ........-.............(((.(((..(((..((.((.((((...((((((((.....)).)----))))))))).)--------------).))..)))..)))..))). ( -26.70) >consensus GGAGACGGCGGCAGAUUGAUGAGGCAAAUUGAUUUGCGGCGCUGCAUCUCUCAAUCUAUUGCAAAA____UUGAGUGCACU_UGCCGCCGUGCUGGGUGAGAAUUGAUUGAGCCA ....(((((((((((((((.((((((((....))))).((...))..))))))))))..((((............))))....))))))))....(((..((.....))..))). (-26.72 = -28.70 + 1.98)

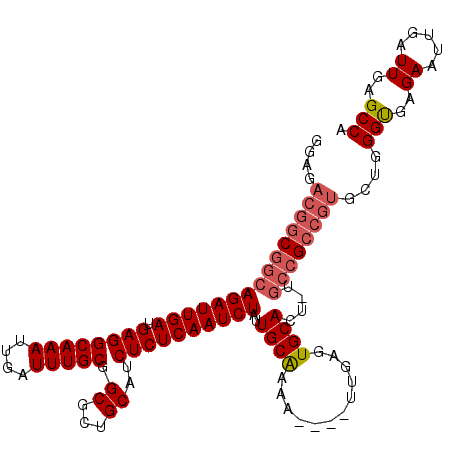
| Location | 5,725,041 – 5,725,156 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.70 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -20.56 |
| Energy contribution | -22.97 |
| Covariance contribution | 2.42 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5725041 115 - 27905053 UGGCUCAAUCAAUUCUCACCCAGCACGGCGGCAUAGUGCACUCAAUCUAUUUUGCAAUAGAUUGAGAGAUGCAGAGCCGCAAAUCAAUUUGCCUCAUCAAUCUGCCGCCGUCUCC .....................((.(((((((((...(((((((((((((((....)))))))))))...))))(((..(((((....)))))))).......))))))))))).. ( -39.60) >DroYak_CAF1 30207 111 - 1 UGGCUCAAUCAAUUCUCACCCAGCACGGCGGCAGAGUGCACUCAA----UUUUGCAAUAGAUUGAGAGAUGCAGCGCCGCAAAUCAAUUUGCCUCAUCAAUCUGCCGCCGUCGCC ......................(((((((((((((.....(((((----(((......)))))))).((((.((....(((((....)))))))))))..))))))))))).)). ( -39.80) >DroAna_CAF1 25535 96 - 1 UGGCCCAAUCAAUUCUCGCC--------------AGUGCACUCAA----UUCCGCAAUAGAUUGAGAGAUGCAGCGCCGCAAAUCAAUUUGCCUCAUCAAUCUCCC-UCAUCUCU .((((..(((.......((.--------------...)).(((((----((........))))))).)))...).)))(((((....)))))..............-........ ( -16.60) >consensus UGGCUCAAUCAAUUCUCACCCAGCACGGCGGCA_AGUGCACUCAA____UUUUGCAAUAGAUUGAGAGAUGCAGCGCCGCAAAUCAAUUUGCCUCAUCAAUCUGCCGCCGUCUCC ........................((((((((....(((..............)))..((((((((((..((...)).(((((....)))))))).))))))))))))))).... (-20.56 = -22.97 + 2.42)

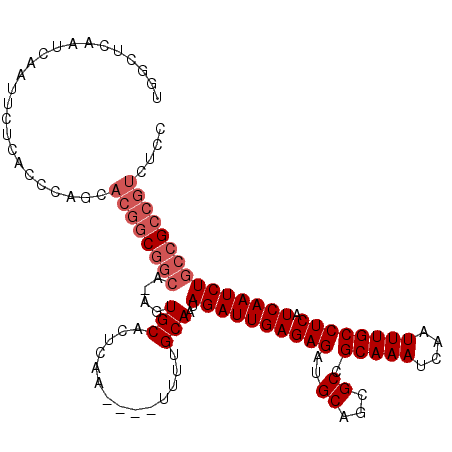

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:19 2006