
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,707,612 – 5,707,722 |
| Length | 110 |
| Max. P | 0.898688 |

| Location | 5,707,612 – 5,707,722 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -15.29 |
| Energy contribution | -14.98 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
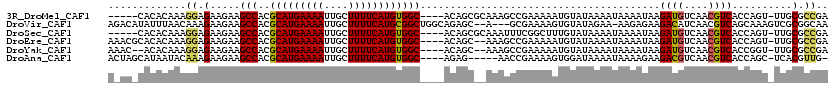
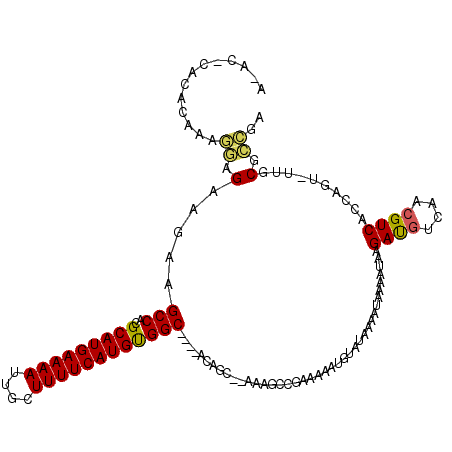
>3R_DroMel_CAF1 5707612 110 + 27905053 -----CACACAAAGGAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC----ACAGCGCAAAGCCGAAAAAUGUAUAAAAUAAAAUAAGAUGUCAACGUCACCAGU-UUGCGCCGA -----.................(((..(((((((((....))))))))))))----.(.(((((((((........))..............((((....)))).....)-)))))).). ( -24.70) >DroVir_CAF1 10108 114 + 1 AGACAUAUUUAACAAAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGCGGCUGGCAGAGC--A---GCGAAAAGUGUAUAGAA-AAGAGAAGACAUCAACGUCAGCAAAGUCGCGGCAA ......................(((.((((((((((....))))))))))((((((...((--(---.(.....))))......-..((.......))...))))))........))).. ( -29.00) >DroSec_CAF1 10906 110 + 1 -----CACACAAAGGAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC----ACAGCGCAAAUUUCGGCUUUGUAUAAAAUAAAAUAAGAUGUCAACGUCACCAGU-UUGCGCCGA -----.................(((..(((((((((....))))))))))))----.(.(((((((((..((.(((((.....)))))....((((....)))).)))))-)))))).). ( -28.40) >DroEre_CAF1 6937 113 + 1 AAACGCACACAAAGGAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC----ACAGC--AAAGCCGAAAAAUGUAUAAAAUAAAAUAAGAUGUCAACGUCACCAGU-UUGCGCCGA ...((((.((...((.......(((..(((((((((....))))))))))))----.....--....)).......................((((....))))....))-.)))).... ( -23.29) >DroYak_CAF1 11249 111 + 1 AAAC--ACACAAAGGAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC----ACAGC--AAAGCCGAAAAAUGUAUAAAAUAAAAUAAGAUGUCAACGUCACCGGU-UUGCGCCGA ....--.......((.......(((..(((((((((....))))))))))))----...((--(((.(((.....(((.....)))......((((....))))..))))-)))).)).. ( -26.20) >DroAna_CAF1 6818 109 + 1 ACUAGCAUAAUACAAAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC----AGAG-----AACCGAAAAGUGGAUAAAAUAAAAGAAGACGUCAACGUCACCAGC-UCACGUUG- ..((((................(((..(((((((((....))))))))))))----.(((-----..(((.....)))..............((((....)))).....)-))..))))- ( -23.30) >consensus A_AC_CACACAAAGGAGAAGAAGCCACGCAUGAAAAUUGCUUUUCAUGUGGC____ACAGC__AAAGCCGAAAAAUGUAUAAAAUAAAAUAAGAUGUCAACGUCACCAGU_UUGCGCCGA .............((.(.....(((..(((((((((....))))))))))))........................................((((....))))..........).)).. (-15.29 = -14.98 + -0.30)
| Location | 5,707,612 – 5,707,722 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.48 |
| Mean single sequence MFE | -28.14 |
| Consensus MFE | -18.08 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
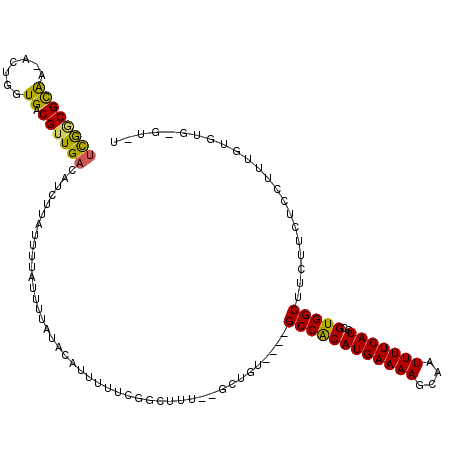
>3R_DroMel_CAF1 5707612 110 - 27905053 UCGGCGCAA-ACUGGUGACGUUGACAUCUUAUUUUAUUUUAUACAUUUUUCGGCUUUGCGCUGU----GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUCCUUUGUGUG----- .((((((((-(..(....)(((((.....(((........)))......)))))))))))))).----((((((((((((....)))))))..))))).................----- ( -29.90) >DroVir_CAF1 10108 114 - 1 UUGCCGCGACUUUGCUGACGUUGAUGUCUUCUCUU-UUCUAUACACUUUUCGC---U--GCUCUGCCAGCCGCAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUUUGUUAAAUAUGUCU ..((((((((...((....))....))).......-...............((---(--(......)))).(((((((((....))))))))))))))...................... ( -28.00) >DroSec_CAF1 10906 110 - 1 UCGGCGCAA-ACUGGUGACGUUGACAUCUUAUUUUAUUUUAUACAAAGCCGAAAUUUGCGCUGU----GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUCCUUUGUGUG----- .((((((((-(.((((....(((....................))).))))...))))))))).----((((((((((((....)))))))..))))).................----- ( -28.95) >DroEre_CAF1 6937 113 - 1 UCGGCGCAA-ACUGGUGACGUUGACAUCUUAUUUUAUUUUAUACAUUUUUCGGCUUU--GCUGU----GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUCCUUUGUGUGCGUUU ..((((((.-((.((.((................................((((...--)))).----((((((((((((....)))))))..)))))....)).))...)).)))))). ( -27.70) >DroYak_CAF1 11249 111 - 1 UCGGCGCAA-ACCGGUGACGUUGACAUCUUAUUUUAUUUUAUACAUUUUUCGGCUUU--GCUGU----GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUCCUUUGUGU--GUUU ...((((((-(.(((..(.(((((.....(((........)))......))))).).--.))).----((((((((((((....)))))))..))))).........)))))))--.... ( -27.70) >DroAna_CAF1 6818 109 - 1 -CAACGUGA-GCUGGUGACGUUGACGUCUUCUUUUAUUUUAUCCACUUUUCGGUU-----CUCU----GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUUUGUAUUAUGCUAGU -........-((((((((((....))))..............((((.....(((.-----....----))).((((((((....)))))))).)))).................)))))) ( -26.60) >consensus UCGGCGCAA_ACUGGUGACGUUGACAUCUUAUUUUAUUUUAUACAUUUUUCGGCUUU__GCUGU____GCCACAUGAAAAGCAAUUUUCAUGCGUGGCUUCUUCUCCUUUGUGUG_GU_U ((((((((.......)).))))))............................................((((((((((((....)))))))..)))))...................... (-18.08 = -17.53 + -0.55)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:11 2006