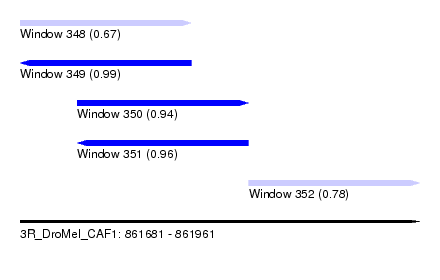
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 861,681 – 861,961 |
| Length | 280 |
| Max. P | 0.994672 |

| Location | 861,681 – 861,801 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -21.90 |
| Energy contribution | -25.52 |
| Covariance contribution | 3.63 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.667146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 861681 120 + 27905053 GUGUGCGGUUUGGCAAACAGGAGCCUUAACUUUGCCGUUAUUCUGGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACUAGCUUCC ((.(((((((.(((........)))..))))...(((.....(((...(((((((.((((((........))))))))))))).)))...)))))).)).....((((((....)))))) ( -38.70) >DroSec_CAF1 36483 120 + 1 GUGUGCGGUUUGGCAAACAGGAACCUUAACUUUGCCGUUAUUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCC ((.(((......))).)).((((((((...((((((............(((((((.((((((........))))))))))))).((....))))))))....(((....))))))))))) ( -39.80) >DroSim_CAF1 35663 120 + 1 GUAUGCGGUUUGGCAAACAGGAACCUUAACUUUGCCGUUAUUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCC ((.(((......))).)).((((((((...((((((............(((((((.((((((........))))))))))))).((....))))))))....(((....))))))))))) ( -39.80) >DroEre_CAF1 36348 93 + 1 GUGUGCGGUUU----------------------GUCGAUAUUCUAUAGGGGCGAAA-----UGGAAUGCAAUUUUAUUUGCUCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUUC .(((((.((((----------------------(((((((((......((((.(((-----((((((...)))))))))))))..)))))).))))))).........)))))....... ( -25.30) >consensus GUGUGCGGUUUGGCAAACAGGAACCUUAACUUUGCCGUUAUUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCC ...................((((((((...((((((............(((((((.((((((........))))))))))))).((....))))))))....(((....))))))))))) (-21.90 = -25.52 + 3.63)

| Location | 861,681 – 861,801 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.47 |
| Mean single sequence MFE | -31.38 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.68 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 861681 120 - 27905053 GGAAGCUAGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCCAGAAUAACGGCAAAGUUAAGGCUCCUGUUUGCCAAACCGCACAC ((((((....))))))..((((((((((((......))))))))))))...............((((((......))))))...((((((....((....)).))))))........... ( -37.80) >DroSec_CAF1 36483 120 - 1 GGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAAUAACGGCAAAGUUAAGGUUCCUGUUUGCCAAACCGCACAC (((((((((.((((((..((((((((((((......))))))))))))...............((((((......))))))...))..))))))))))))).((.(((......))).)) ( -36.00) >DroSim_CAF1 35663 120 - 1 GGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAAUAACGGCAAAGUUAAGGUUCCUGUUUGCCAAACCGCAUAC (((((((((.((((((..((((((((((((......))))))))))))...............((((((......))))))...))..))))))))))))).((.(((......))).)) ( -36.20) >DroEre_CAF1 36348 93 - 1 GAAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGAGCAAAUAAAAUUGCAUUCCA-----UUUCGCCCCUAUAGAAUAUCGAC----------------------AAACCGCACAC .......(((((......((((((((((((....)).).)))))))))...........-----.(((.........))).......----------------------.....))))). ( -15.50) >consensus GGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAAUAACGGCAAAGUUAAGGUUCCUGUUUGCCAAACCGCACAC ........((((......((((((((((((......))))))))))))...............((((((......))))))...((((((....((....)).)))))).....)))).. (-22.42 = -23.68 + 1.25)

| Location | 861,721 – 861,841 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 861721 120 + 27905053 UUCUGGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACUAGCUUCCAGCCCUUUUCUUGUCUUUUGUCACAGGCACAUUCGACAAA ....((((((((.(((((((((........)))))))))((((........))))........(((((((....))))))))))))))).(((((...((((....).)))...))))). ( -38.50) >DroSec_CAF1 36523 120 + 1 UUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCCAGCCCUUUUCUUGUCUUUUGUGACAGGCACAUUUGACAAA ....((((((((....((((((..........(((.(((((((........))))))).)))(((....)))...)))))))))))))).(((((...((((.....))))...))))). ( -39.10) >DroSim_CAF1 35703 120 + 1 UUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCCAGCCCUUUUCUUGUCUUUUGUCACAGGCACAUUUGACAAA .....(((((((((...((((.((..(((((((((.(((((((........)))))))....(((....))))))))))))..)).)))))))))))))((((.(......).))))... ( -37.30) >DroEre_CAF1 36366 115 + 1 UUCUAUAGGGGCGAAA-----UGGAAUGCAAUUUUAUUUGCUCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUUCAGCCCUUUUCUUGUCUUUUGUCACAGGAGCAUUUGACAAA ......((((((((((-----(....(((.(((((.(((((((........))))))).)))))....)))....))))).))))))...(((((...(((.......)))...))))). ( -30.10) >consensus UUCUAGAGGGGCAGAAUGGAAUGGAAUGGAAUUUUAUUUGCCCACGGUAUCGGGCAAACAAAGUGGAAGCACAAGGUUCCAGCCCUUUUCUUGUCUUUUGUCACAGGCACAUUUGACAAA ....((((((((..................(((((.(((((((........))))))).)))))((((.(....).)))).)))))))).(((((...(((.......)))...))))). (-32.70 = -32.20 + -0.50)

| Location | 861,721 – 861,841 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -28.09 |
| Energy contribution | -28.02 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959658 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 861721 120 - 27905053 UUUGUCGAAUGUGCCUGUGACAAAAGACAAGAAAAGGGCUGGAAGCUAGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCCAGAA ((((((...(((.......)))...))))))....(((((((((((....))))))).((((((((((((......))))))))))))....................))))........ ( -39.10) >DroSec_CAF1 36523 120 - 1 UUUGUCAAAUGUGCCUGUCACAAAAGACAAGAAAAGGGCUGGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAA ((((((...((((.....))))...))))))....(((((((((....(((....)))((((((((((((......))))))))))))...........)))))....))))........ ( -33.80) >DroSim_CAF1 35703 120 - 1 UUUGUCAAAUGUGCCUGUGACAAAAGACAAGAAAAGGGCUGGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAA (((((((..........))))))).....(((...(((((((((....(((....)))((((((((((((......))))))))))))...........)))))....)))).))).... ( -32.00) >DroEre_CAF1 36366 115 - 1 UUUGUCAAAUGCUCCUGUGACAAAAGACAAGAAAAGGGCUGAAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGAGCAAAUAAAAUUGCAUUCCA-----UUUCGCCCCUAUAGAA (((((((..........)))))))...........((((.((((....((((......((((((((((((....)).).)))))))))...))))....-----))))))))........ ( -27.30) >consensus UUUGUCAAAUGUGCCUGUGACAAAAGACAAGAAAAGGGCUGGAACCUUGUGCUUCCACUUUGUUUGCCCGAUACCGUGGGCAAAUAAAAUUCCAUUCCAUUCCAUUCUGCCCCUCUAGAA ((((((...(((.......)))...))))))....((((.((((....(((....)))((((((((((((......))))))))))))................))))))))........ (-28.09 = -28.02 + -0.06)

| Location | 861,841 – 861,961 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.90 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -29.84 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.780922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 861841 120 + 27905053 CAGUCAGCGCGAAGUGGGAACUGGGAACUGGUAUAGAGCUUCCCAGCCAAGUUCAAUUGGGUUGACUUCGGCGGCAGCUGAUUUCGAUUUGUAUCUAAUUGUAUGCAAACAAAAUCACAU ..(((.((.((((((.((..(((((((((.......)).)))))))((((......)))).)).))))))))))).(.((((((.(.((((((((.....).)))))))).))))))).. ( -36.50) >DroSec_CAF1 36643 118 + 1 CAGUCAGC--GAAGUGGGAACUGGGAACUGGGAUAGAGGUUCCCAGCCAAGUUCAAUUGGGUUGACUUUGGCGGCAGCUGAUUCCGAUUUGUAUCUAAUUGUAUGCAAACAAAAUCACAU .(((((((--...........((((((((........))))))))((((((.(((((...))))).))))))....)))))))..(.((((((((.....).)))))))).......... ( -36.10) >DroSim_CAF1 35823 118 + 1 CAGUCAGC--GAAGUGGGAACUGGGAACUGGGAUAGAGUUUCCCAGCCAAGUUCAAUUGGGUUGACUUCGGCGGCAGCUGAUUCCGAUUUGUAUCUAAUUGUAUGCAAACAAAAUCACAU .(((((((--..(((..(((((.((..((((((.......)))))))).))))).)))..)))))))(((((....)))))....(.((((((((.....).)))))))).......... ( -37.40) >DroEre_CAF1 36481 109 + 1 CAGUCGGC--GAG---------CGGAACUGGGAUAGAGUUUCCCAGCCAGGUUCAACUGGGUUGACUUCGGCGGCAGCUGAUUCCGAUUUGUAUCUAAUUGUAUGCAAACAGAAUCACAU ..((((.(--(((---------.(...((((((.......))))))((((......)))).....))))).)))).(.((((((.(.((((((((.....).)))))))).))))))).. ( -37.30) >consensus CAGUCAGC__GAAGUGGGAACUGGGAACUGGGAUAGAGUUUCCCAGCCAAGUUCAAUUGGGUUGACUUCGGCGGCAGCUGAUUCCGAUUUGUAUCUAAUUGUAUGCAAACAAAAUCACAU .(((((((...................((((((.......))))))((((......)))))))))))..(((....)))(((((.(.((((((((.....).)))))))).))))).... (-29.84 = -29.90 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:16 2006