
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,699,804 – 5,700,029 |
| Length | 225 |
| Max. P | 0.968389 |

| Location | 5,699,804 – 5,699,918 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -30.37 |
| Consensus MFE | -28.29 |
| Energy contribution | -28.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5699804 114 - 27905053 UUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAACAUCAGUCUCAAAACAAACGA--GAGAGAAAGAGCG .................((..(((((((((((...(....)((((((....))))))...)))))))))))...........((..((((...........--..))))..)))). ( -29.52) >DroSim_CAF1 2654 116 - 1 UUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAGCAGCGAACAUCAGUCUCACAACAAACGAGAGAGAGAAAGAGCG .................((..(((((((((((...(....)((((((....))))))...))))))))))).)).((.....((..((((.........))))..))......)). ( -32.80) >DroYak_CAF1 3064 113 - 1 UUAUCAAUAACAACAGAGCUAUAACAGCUGUUGGAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAACAUCAGCCCCAAAACAAACGAGAGAGAGAGAGA--- ...((...........((((.....)))).((((.(((..(((((((....)))))))......((((.....))))........))))))).......))............--- ( -28.80) >consensus UUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGCAACAAUGUUGCGAAAACAGCUGUUGAACAGCGAACAUCAGUCUCAAAACAAACGAGAGAGAGAAAGAGCG ...((.....((((((...........))))))(((((..(((((((....)))))))......((((.....))))........)))))...............))......... (-28.29 = -28.40 + 0.11)

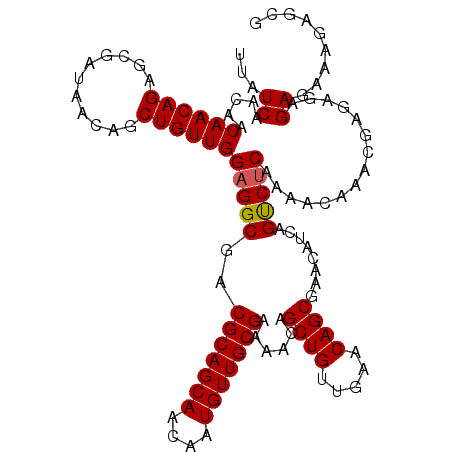

| Location | 5,699,840 – 5,699,950 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.09 |
| Mean single sequence MFE | -35.89 |
| Consensus MFE | -17.78 |
| Energy contribution | -19.83 |
| Covariance contribution | 2.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884915 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5699840 110 - 27905053 AAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGC-AACAAUGUUGCGAAAACAGCUG-UUGAACAGCG .....((.(--------((...........((((((((...............))))))))(((((((((((...(....)(((((-(....))))))...))))))))-))).))))). ( -39.26) >DroPse_CAF1 16137 100 - 1 ACUGUGCGUGUAUCAGUGUGUGUGUGCGUUUCGCUCUGUAUUAUCAAUA---ACAGAGCGGA-----------GAGGAGA-ACAGCGAACA-----GCGAACAGAGUGGAACGAGCAGCG .((((.(((...(((.(.(((.(((.(.(((((((((((..........---))))))))))-----------)..)...-)))((.....-----))..))).).))).))).)))).. ( -30.70) >DroSec_CAF1 3185 110 - 1 AAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGC-AACAAUGUUGCGAAAACAGCUG-GUGAACAGCG ..((..(..--------......)..))..((((((((...............)))))))).....((((((.(.(((..((((((-(....)))))))......))).-.).)))))). ( -37.26) >DroSim_CAF1 2692 110 - 1 AAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGC-AACAAUGUUGCGAAAACAGCUG-UUGAGCAGCG .....((.(--------((...........((((((((...............))))))))(((((((((((...(....)(((((-(....))))))...))))))))-))).))))). ( -39.56) >DroYak_CAF1 3099 110 - 1 AAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGUUUAUCAAUAACAACAGAGCUAUAACAGCUGUUGGAGGCGACGCAGC-AACAAUGUUGCGAAAACAGCUG-UUGAACAGCG .....((.(--------((..(((.....)))((((((...............))))))..(((((((((((...(....)(((((-(....))))))...))))))))-))).))))). ( -37.86) >DroPer_CAF1 18514 100 - 1 ACUGUGCGUGUAUCAGUGUGUGUGUGCGUUUCGCUCUGUAUUAUCAAUA---ACAGAGCGGA-----------GAGGAGA-ACAGCGAACA-----GCGAACAGAGUGGAACGAGCAGCG .((((.(((...(((.(.(((.(((.(.(((((((((((..........---))))))))))-----------)..)...-)))((.....-----))..))).).))).))).)))).. ( -30.70) >consensus AAUGUGCGU________GUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGACGCAGC_AACAAUGUUGCGAAAACAGCUG_UUGAACAGCG ..(((.(((.........(((...((((..((((((((...............)))))))).....(((......)))..))))....))).....)))...)))((((......)))). (-17.78 = -19.83 + 2.06)

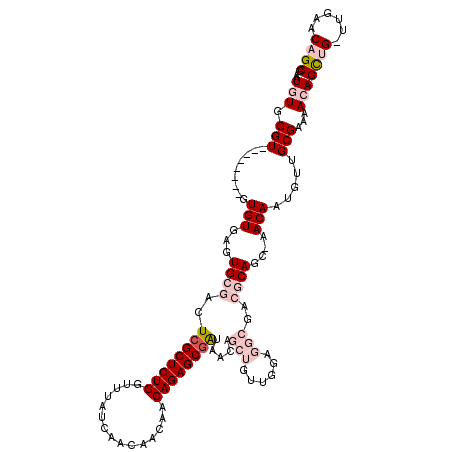

| Location | 5,699,878 – 5,699,989 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 97.31 |
| Mean single sequence MFE | -41.81 |
| Consensus MFE | -40.19 |
| Energy contribution | -40.69 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5699878 111 - 27905053 CAAAUCUACAGAGCACUUGUC-ACGCAUACGCGCAUCAUAAAUGUGCGUGUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGA ....(((((((.(((((....-.(((((((((((((.....))))))))))))))))))...((((((((...............))))))))......))).))))..... ( -43.26) >DroSec_CAF1 3223 111 - 1 UA-AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGUGUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGA ..-.(((((((.(((((......(((((((((((((.....))))))))))))))))))...((((((((...............))))))))......))).))))..... ( -42.86) >DroSim_CAF1 2730 111 - 1 CA-AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGUGUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGA ..-.(((((((.(((((......(((((((((((((.....))))))))))))))))))...((((((((...............))))))))......))).))))..... ( -42.86) >DroYak_CAF1 3137 111 - 1 CA-AUCUACAUAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGUGUGUGAGUGCGACUCGCUCUGGUUUAUCAAUAACAACAGAGCUAUAACAGCUGUUGGAGGCGA ..-.........(((((......(((((((((((((.....))))))))))))))))))..(((((((((...............))))))..((((....))))))).... ( -38.26) >consensus CA_AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGUGUGUGAGUGCGACUCGCUCUGGUUUAUCAACAACAACAGAGCGAUAACAGCUGUUGGAGGCGA ....(((((((.(((((......(((((((((((((.....))))))))))))))))))...((((((((...............))))))))......))).))))..... (-40.19 = -40.69 + 0.50)



| Location | 5,699,918 – 5,700,029 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.91 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -13.14 |
| Energy contribution | -15.75 |
| Covariance contribution | 2.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5699918 111 - 27905053 AAGAGCAGCAUAAAAUACUACGCAGUUAAAAAAACAAAACCAAAUCUACAGAGCACUUGUC-ACGCAUACGCGCAUCAUAAAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGU .(((((.((.((......)).))((((.................((....))(((((....-.(((((((((((((.....))))))))--------)))))))))))))).)))))... ( -34.30) >DroPse_CAF1 16197 102 - 1 -AAAAU----AAAAAUACUACGCAGUAAAAAAA--AAAACU-------CAGA-CACUUGUCAACAGAGUCCCGC---AUAACUGUGCGUGUAUCAGUGUGUGUGUGCGUUUCGCUCUGUA -.....----.....((((....))))......--......-------..((-(....))).(((((((..(((---(((...(..(........)..)...))))))....))))))). ( -21.90) >DroSec_CAF1 3263 111 - 1 AAGAGCAGCAUAAAAUACUACGCAGUUAAAAAAACAAAACUA-AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGU .(((((.((.((......)).))((((...............-.((....))(((((......(((((((((((((.....))))))))--------)))))))))))))).)))))... ( -33.90) >DroSim_CAF1 2770 111 - 1 AAGAGCAGCAUAAAAUACUACGCAGUUAAAAAAACAAAACCA-AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGU .(((((.((.((......)).))((((...............-.((....))(((((......(((((((((((((.....))))))))--------)))))))))))))).)))))... ( -33.90) >DroYak_CAF1 3177 111 - 1 AAGAGCAGCAUAAAAUACUACGCAGUUAUGAAAACAAAACCA-AUCUACAUAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGU--------GUGUGAGUGCGACUCGCUCUGGU .(((((((.........)).((((((((((............-.....)))))).........(((((((((((((.....))))))))--------)))))..))))....)))))... ( -33.83) >DroPer_CAF1 18574 102 - 1 AAAAAU----AAAAAUACUACGCAGUAAAAA-A--AAAACU-------CAGA-CACUUGUCAACAGAGUCCCGC---AUAACUGUGCGUGUAUCAGUGUGUGUGUGCGUUUCGCUCUGUA ......----.....((((....))))....-.--......-------..((-(....))).(((((((..(((---(((...(..(........)..)...))))))....))))))). ( -21.90) >consensus AAGAGCAGCAUAAAAUACUACGCAGUUAAAAAAACAAAACCA_AUCUACAGAGCACUUGUCAACGCAUACGCGCAUCAUAAAUGUGCGU________GUGUGAGUGCGACUCGCUCUGGU ................................................((((((..((((...(((((((((((((.....))))))))........)))))...))))...)))))).. (-13.14 = -15.75 + 2.61)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:25:03 2006