
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,678,294 – 5,678,426 |
| Length | 132 |
| Max. P | 0.978161 |

| Location | 5,678,294 – 5,678,393 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -38.44 |
| Consensus MFE | -24.44 |
| Energy contribution | -26.00 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5678294 99 + 27905053 AG-GGGUUGGUUUUCCGCCGUAAUCGCAGUUCCUGCAGGUGGCUACUUAGGGAGUUUCCGUGAAUUCACGGGAAAAACUGCAGUGGCCAAGUUGG------GAAUA .(-.(((((((.....)))..)))).).((((((.....(((((((((((....((((((((....))))))))...))).))))))))....))------)))). ( -37.60) >DroSec_CAF1 19689 98 + 1 AG-GGGUGGGUUUUCCGCCGUAAUCGCAGUUCCUGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGAUGA-AAACUGCAGUGGCCAAGGUGG------GAAUA ..-.(((((.....))))).........(((((..(...(((((((((((((.....)))))).(((((.(..-..)))))))))))))..)..)------)))). ( -44.70) >DroSim_CAF1 19804 98 + 1 AG-GGGUGGGUUUUCCGCCGUAAUCGCAGUUCCUGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGCUGA-AAACUGCAGUGGCCAAGUUGG------GAAUA ..-.(((((.....))))).........((((((.....(((((((((((((.....)))))).(((((.(..-..)))))))))))))....))------)))). ( -40.00) >DroEre_CAF1 16767 85 + 1 GGUGGGUGGGUUUUCCGCAGUAAUCGCGGUUCCUGCAGGUGGCUACUUAGGGAGUUUUCCUGAAUGCAGGGAA-AACCUGCAGUG--------------------A .....(..((((((((((((.(((....))).)))).....((...(((((((...)))))))..))..))))-))))..)....--------------------. ( -31.10) >DroYak_CAF1 20332 105 + 1 AGUGGUUGGGUUUUCCGCUGUAAUCGCAGUUCCUGCAGGUGGCUACUUAGGGAGUUUUCCCGAAUGCAGGGCA-AAAUUGCAGUGGCCAAGGUGGGAAUGGGAAUA (((((.........)))))....((.((.((((..(...(((((((...(((......)))...(((((....-...))))))))))))..)..)))))).))... ( -38.80) >consensus AG_GGGUGGGUUUUCCGCCGUAAUCGCAGUUCCUGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGGGGA_AAACUGCAGUGGCCAAGGUGG______GAAUA ....(((((.....)))))......((((...))))...(((((((((((((.....)))))).(((((........))))))))))))................. (-24.44 = -26.00 + 1.56)

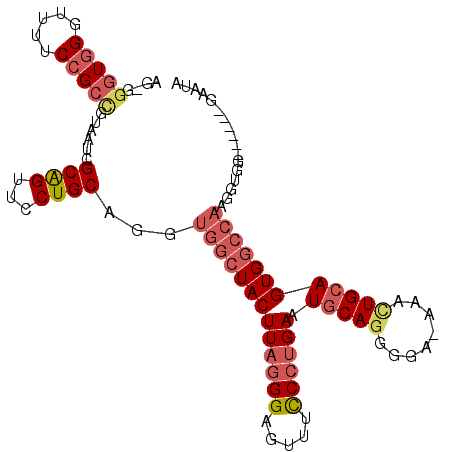
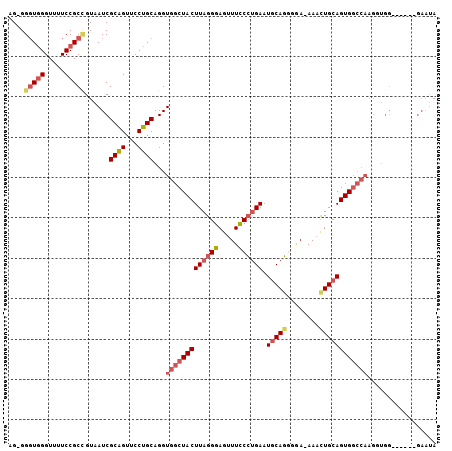
| Location | 5,678,294 – 5,678,393 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -26.10 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5678294 99 - 27905053 UAUUC------CCAACUUGGCCACUGCAGUUUUUCCCGUGAAUUCACGGAAACUCCCUAAGUAGCCACCUGCAGGAACUGCGAUUACGGCGGAAAACCAACCC-CU ..(((------((.....)(((..((((((((((.(((((....)))))...........((((....)))))))))))))).....))))))).........-.. ( -25.80) >DroSec_CAF1 19689 98 - 1 UAUUC------CCACCUUGGCCACUGCAGUUU-UCAUCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCAGGAACUGCGAUUACGGCGGAAAACCCACCC-CU .....------((.((.((((.(((((((...-....))))....((((.....)))).))).))))..(((((...))))).....)).))...........-.. ( -27.70) >DroSim_CAF1 19804 98 - 1 UAUUC------CCAACUUGGCCACUGCAGUUU-UCAGCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCAGGAACUGCGAUUACGGCGGAAAACCCACCC-CU .....------......((((.((((((((..-...)))))....((((.....)))).))).))))..(((((...))))).....((.((.....)).)).-.. ( -28.30) >DroEre_CAF1 16767 85 - 1 U--------------------CACUGCAGGUU-UUCCCUGCAUUCAGGAAAACUCCCUAAGUAGCCACCUGCAGGAACCGCGAUUACUGCGGAAAACCCACCCACC (--------------------(.((((((((.-....((((.((..(((....)))..))))))..)))))))))).(((((.....))))).............. ( -25.90) >DroYak_CAF1 20332 105 - 1 UAUUCCCAUUCCCACCUUGGCCACUGCAAUUU-UGCCCUGCAUUCGGGAAAACUCCCUAAGUAGCCACCUGCAGGAACUGCGAUUACAGCGGAAAACCCAACCACU ..((((..((((.....((((.((((((....-.....)))....((((....))))..))).))))......))))..((.......))))))............ ( -22.80) >consensus UAUUC______CCAACUUGGCCACUGCAGUUU_UCCCCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCAGGAACUGCGAUUACGGCGGAAAACCCACCC_CU .......................((((((.......((((....))))...(((.....)))......))))))...((((.......)))).............. (-16.06 = -15.70 + -0.36)


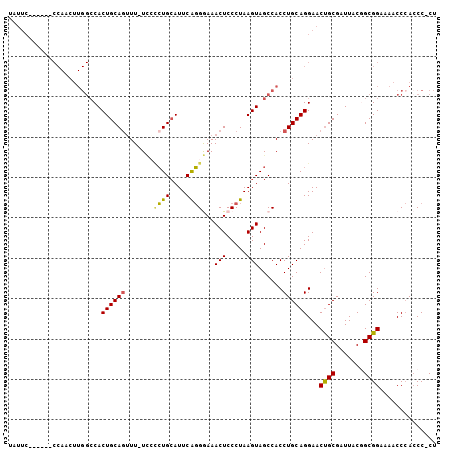
| Location | 5,678,326 – 5,678,426 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 85.60 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -19.80 |
| Energy contribution | -21.55 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5678326 100 + 27905053 UGCAGGUGGCUACUUAGGGAGUUUCCGUGAAUUCACGGGAAAAACUGCAGUGGCCAAGUUGG------GAAUACAAUAAAGAAAUGGGU-UUGAAAAAGGGAAAGCU .((...(((((((((((....((((((((....))))))))...))).)))))))).((((.------.....))))............-..............)). ( -30.10) >DroSec_CAF1 19721 99 + 1 UGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGAUGA-AAACUGCAGUGGCCAAGGUGG------GAAUACAAUAAAGAAAUGAGU-CUGAAAAGGGAAAAGCU .((...(((((((((((((.....)))))).(((((.(..-..)))))))))))))......------....................(-((....))).....)). ( -28.60) >DroSim_CAF1 19836 99 + 1 UGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGCUGA-AAACUGCAGUGGCCAAGUUGG------GAAUACAAUAAAGAAAUGGGU-CUGAAAAGGGAAAAGCU .((...(((((((((((((.....)))))).(((((.(..-..))))))))))))).((((.------.....))))...........(-((....))).....)). ( -30.10) >DroYak_CAF1 20365 106 + 1 UGCAGGUGGCUACUUAGGGAGUUUUCCCGAAUGCAGGGCA-AAAUUGCAGUGGCCAAGGUGGGAAUGGGAAUACAAAAGGGAAAUGGCUGCUGAAAGGGGGAUAGCU .((...(((((((...(((......)))...(((((....-...))))))))))))...............................((.((....)).))...)). ( -26.00) >consensus UGCAGGUGGCUACUUAGGGAGUUUCCCUGAAUGCAGGGGA_AAACUGCAGUGGCCAAGGUGG______GAAUACAAUAAAGAAAUGGGU_CUGAAAAGGGAAAAGCU .((...(((((((((((((.....)))))).(((((........))))))))))))..................................((....))......)). (-19.80 = -21.55 + 1.75)

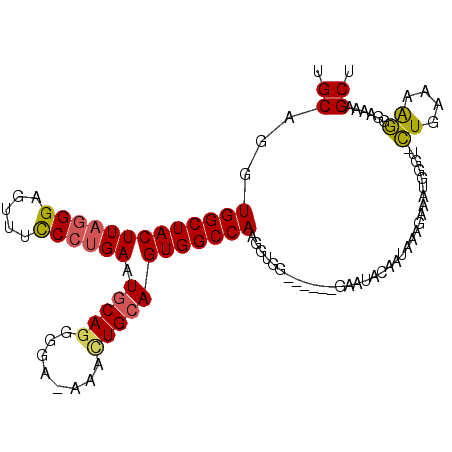

| Location | 5,678,326 – 5,678,426 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 85.60 |
| Mean single sequence MFE | -19.47 |
| Consensus MFE | -12.21 |
| Energy contribution | -13.28 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.785480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5678326 100 - 27905053 AGCUUUCCCUUUUUCAA-ACCCAUUUCUUUAUUGUAUUC------CCAACUUGGCCACUGCAGUUUUUCCCGUGAAUUCACGGAAACUCCCUAAGUAGCCACCUGCA .((..............-.............(((.....------.)))..((((.(((..(((((...(((((....)))))))))).....))).))))...)). ( -17.90) >DroSec_CAF1 19721 99 - 1 AGCUUUUCCCUUUUCAG-ACUCAUUUCUUUAUUGUAUUC------CCACCUUGGCCACUGCAGUUU-UCAUCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCA .((............((-(......)))...........------......((((.(((((((...-....))))....((((.....)))).))).))))...)). ( -19.80) >DroSim_CAF1 19836 99 - 1 AGCUUUUCCCUUUUCAG-ACCCAUUUCUUUAUUGUAUUC------CCAACUUGGCCACUGCAGUUU-UCAGCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCA .((............((-(......)))...........------......((((.((((((((..-...)))))....((((.....)))).))).))))...)). ( -21.70) >DroYak_CAF1 20365 106 - 1 AGCUAUCCCCCUUUCAGCAGCCAUUUCCCUUUUGUAUUCCCAUUCCCACCUUGGCCACUGCAAUUU-UGCCCUGCAUUCGGGAAAACUCCCUAAGUAGCCACCUGCA .(((((..........(((((((.........((......)).........))))....(((....-)))..)))....((((....))))...)))))........ ( -18.47) >consensus AGCUUUCCCCUUUUCAG_ACCCAUUUCUUUAUUGUAUUC______CCAACUUGGCCACUGCAGUUU_UCACCUGCAUUCAGGGAAACUCCCUAAGUAGCCACCUGCA ...................................................((((.(((((((........))))....((((.....)))).))).))))...... (-12.21 = -13.28 + 1.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:54 2006