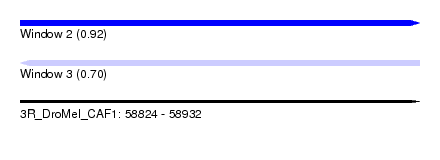
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 58,824 – 58,932 |
| Length | 108 |
| Max. P | 0.919711 |

| Location | 58,824 – 58,932 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -21.79 |
| Energy contribution | -22.93 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
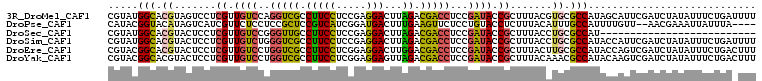
>3R_DroMel_CAF1 58824 108 + 27905053 AAAAUCAGAAAUAUAGAUCGAAUGCUAUGGCGCACGUAAAGCGGUAUCGGAGGUCGUCUAAGUCCUCGGAGGAAGGCGACCUGGACAACGAGGACUACGUGCCAUACG .........................((((((((.((.....))((.(((.(((((((((...(((.....))))))))))))......)))..))...)))))))).. ( -31.50) >DroPse_CAF1 1926 102 + 1 ----UAAAUAAUUUCGUU--AACAAAAUGGCAAAUGUAAAGAGGUACAGGAGAACUUCAAAGUCAUCCGAUGACGGAGCGGAGGAGAACGAUGACUAUGUACCGUAUG ----...........(((--(......))))...........(((((((((....)))..(((((((((....)))).((........)).))))).))))))..... ( -20.00) >DroSec_CAF1 1 82 + 1 --------------------------AUGGCGCAGGUAAAGCGGUAUCGGAGGUCGUCUAAGUCCUCGGAGGAAGGCAACCCGGACAACGAGGAGUACGUGCCAUACG --------------------------(((((((.......((..(.((.((((.(......).)))).))..)..))..((((.....)).)).....)))))))... ( -27.00) >DroSim_CAF1 3854 108 + 1 AAAAUCAGAAAUAUAGAUCGAAUGGUAUGGCGCAGGUAAAGCGGUAUCGGAGGUCGUCUAAGUCCUCGGAGGAAGGCGACCCAGACAACGAGGAGUACGUGCCAUACG ........................(((((((((..(((...((...((.(.((((((((...(((.....)))))))))))).))...)).....)))))))))))). ( -34.20) >DroEre_CAF1 1976 108 + 1 AAAGUCAGAAAUAUAGAUCGACUGGUAUGGCGCAAGUAAAGCGGUAUCGGAGGUCGUCCAAGUCCUCCGAGGAAGGCGACCAGGACAACGAGGAGUACGUGCCGUACG ..((((.((........)))))).(((((((((.......((..(.(((((((.(......).)))))))..)..))..((.(.....)..)).....))))))))). ( -35.90) >DroYak_CAF1 3841 108 + 1 AAAGUCAGAAAUAUAGAUCGACUUGUAUGGCGUUUGUAAAGCGGUAUCGGAGGUCGUCUAACUCCUCCGAGGAAGGCGACCAGGACAACGAGGAGUACGUGCCGUACG .(((((.((........)))))))((((((((..((((...((...((...((((((((..(((....)))..))))))))..))...)).....)))))))))))). ( -36.20) >consensus AAA_UCAGAAAUAUAGAUCGAAUGGUAUGGCGCAGGUAAAGCGGUAUCGGAGGUCGUCUAAGUCCUCCGAGGAAGGCGACCAGGACAACGAGGAGUACGUGCCAUACG ........................(((((((((..(((...((...((...((((((((...(((.....)))))))))))..))...)).....)))))))))))). (-21.79 = -22.93 + 1.14)


| Location | 58,824 – 58,932 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.02 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -16.20 |
| Energy contribution | -15.79 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.700127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 58824 108 - 27905053 CGUAUGGCACGUAGUCCUCGUUGUCCAGGUCGCCUUCCUCCGAGGACUUAGACGACCUCCGAUACCGCUUUACGUGCGCCAUAGCAUUCGAUCUAUAUUUCUGAUUUU ..((((((((((((....((.((((.((((((.(((((.....)))...)).))))))..)))).))..))))))..))))))......((((.........)))).. ( -28.90) >DroPse_CAF1 1926 102 - 1 CAUACGGUACAUAGUCAUCGUUCUCCUCCGCUCCGUCAUCGGAUGACUUUGAAGUUCUCCUGUACCUCUUUACAUUUGCCAUUUUGUU--AACGAAAUUAUUUA---- .....((((((.(((((((((........))..((....))))))))).)).........((((......))))..))))((((((..--..))))))......---- ( -16.80) >DroSec_CAF1 1 82 - 1 CGUAUGGCACGUACUCCUCGUUGUCCGGGUUGCCUUCCUCCGAGGACUUAGACGACCUCCGAUACCGCUUUACCUGCGCCAU-------------------------- ...(((((..(((.((...((((((..(((..((((.....)))))))..))))))....))))).((.......)))))))-------------------------- ( -22.70) >DroSim_CAF1 3854 108 - 1 CGUAUGGCACGUACUCCUCGUUGUCUGGGUCGCCUUCCUCCGAGGACUUAGACGACCUCCGAUACCGCUUUACCUGCGCCAUACCAUUCGAUCUAUAUUUCUGAUUUU .(((((((..(((.((...(((((((((((..((((.....)))))))))))))))....))))).((.......))))))))).....((((.........)))).. ( -30.60) >DroEre_CAF1 1976 108 - 1 CGUACGGCACGUACUCCUCGUUGUCCUGGUCGCCUUCCUCGGAGGACUUGGACGACCUCCGAUACCGCUUUACUUGCGCCAUACCAGUCGAUCUAUAUUUCUGACUUU .(((.(((..(((.((...(((((((.((((.((......))..)))).)))))))....))))).((.......))))).))).((((((........)).)))).. ( -30.70) >DroYak_CAF1 3841 108 - 1 CGUACGGCACGUACUCCUCGUUGUCCUGGUCGCCUUCCUCGGAGGAGUUAGACGACCUCCGAUACCGCUUUACAAACGCCAUACAAGUCGAUCUAUAUUUCUGACUUU .(((.(((..........((.((((..(((((.((..(((....)))..)).)))))...)))).))..........))).)))(((((((........)).))))). ( -28.85) >consensus CGUACGGCACGUACUCCUCGUUGUCCUGGUCGCCUUCCUCCGAGGACUUAGACGACCUCCGAUACCGCUUUACAUGCGCCAUACCAUUCGAUCUAUAUUUCUGA_UUU .....(((.((.......((.((((..(((((.(((((.....)))...)).)))))...)))).)).......)).)))............................ (-16.20 = -15.79 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:29:47 2006