
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,640,254 – 5,640,445 |
| Length | 191 |
| Max. P | 0.994366 |

| Location | 5,640,254 – 5,640,374 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.81 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -28.59 |
| Energy contribution | -28.78 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5640254 120 - 27905053 CAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUUUUGCGGAGCACAAAGAAAAUGGUUUCCACAAGGUCUACGAAUUUGUUCACCAGGUACAAAGUGCAAAUCAAACGCU .....((((((.((((..((((((((((((......))))))))))))((((..((.......))..)))).....))))..(((....))).))..((((...))))..))))...... ( -30.00) >DroSec_CAF1 14081 120 - 1 CAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACGAAGAAUAUCGUUUCCACAAGGUCCACGAAUUUGUUCAGCAUGUACAAAAUGCAAAUCAAACGCU .....(((((((....((((((((((((((......))))))))))))((((.((((......))))))))))....))).............((((.......))))..))))...... ( -35.70) >DroSim_CAF1 14785 120 - 1 CAAAAUGGUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACGAAGAAAAUCGUUUCCACAAGGUCUACGAAUUUGUUCAGCAUGUACAAAAUGCAAAUCAAACGCU ......((((..((((..((((((((((((......))))))))))))((((.((((......)))))))).....)))).....((((....((((.......))))....)))))))) ( -37.70) >DroYak_CAF1 14983 120 - 1 AAAAGUGAUGAAAGACGUGCAGGAAGGAUAUCUGUUUGUCCUUCUUGCGGAGCACAAAGAAAACCGUUUCCACAAGGUCCACGAAUUUGUUAAACAUAUACAAAAUGCAUAUCGAACGCU ...((((........(((((((((((((((......))))))))))))(((.(.....((((....)))).....).))))))..(((((.........)))))............)))) ( -30.20) >consensus CAAAAUGAUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACAAAGAAAAUCGUUUCCACAAGGUCCACGAAUUUGUUCAGCAUGUACAAAAUGCAAAUCAAACGCU .....((((....(((..((((((((((((......))))))))))))((((.((((......)))))))).....)))......(((((.........)))))......))))...... (-28.59 = -28.78 + 0.19)

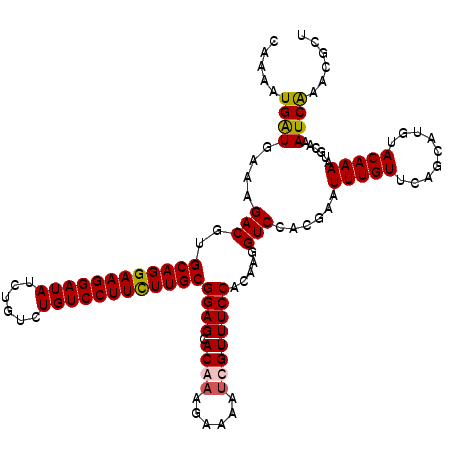
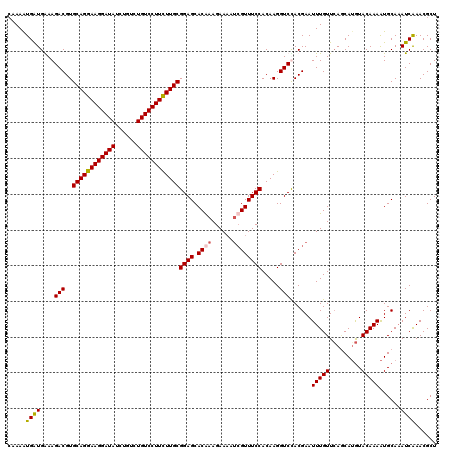
| Location | 5,640,294 – 5,640,414 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -35.84 |
| Consensus MFE | -32.68 |
| Energy contribution | -32.86 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5640294 120 + 27905053 AGACCUUGUGGAAACCAUUUUCUUUGUGCUCCGCAAAAAGGACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUUUUGGAUAUCUCUACUACUGCUUCCACGG ......(((((((......((((((.(((...))).))))))(((..((((((((....(((.((((...((......)).)))))))......))))))))......))).))))))). ( -29.20) >DroSec_CAF1 14121 120 + 1 GGACCUUGUGGAAACGAUAUUCUUCGUGCUCCGCAAGAAGGACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUCUGGGAUAUCUGUACUACUACUUCCACGG (((.((((((((.((((......))))..))))))))......(((((((((((((...(((.((((...((......)).))))))).....))))))))))).))......))).... ( -42.30) >DroSim_CAF1 14825 120 + 1 AGACCUUGUGGAAACGAUUUUCUUCGUGCUCCGCAAGAAGGACAGACAGAUAUCCUUCCUGCACGUCUUUCACCAUUUUGCGAUGGCAUUUCUGGGAUAUCUCUACUACUACUUCCACGG ...(((((((((.((((......))))..)))))))((((...((..(((((((((...(((.((((..............))))))).....)))))))))...))....))))...)) ( -34.04) >DroYak_CAF1 15023 120 + 1 GGACCUUGUGGAAACGGUUUUCUUUGUGCUCCGCAAGAAGGACAAACAGAUAUCCUUCCUGCACGUCUUUCAUCACUUUUCGAUGGCAUUUUUCGGAUAUCUGUAUUACUUUUUCCACGG ......((((((((.(((.....((((.((.(....).)).))))((((((((((....(((.((((..............)))))))......))))))))))...))).)))))))). ( -37.84) >consensus AGACCUUGUGGAAACGAUUUUCUUCGUGCUCCGCAAGAAGGACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUCUGGGAUAUCUCUACUACUACUUCCACGG ......(((((((......((((((.(((...))).)))))).(((((((((((((...(((.((((..............))))))).....))))))))))).)).....))))))). (-32.68 = -32.86 + 0.19)

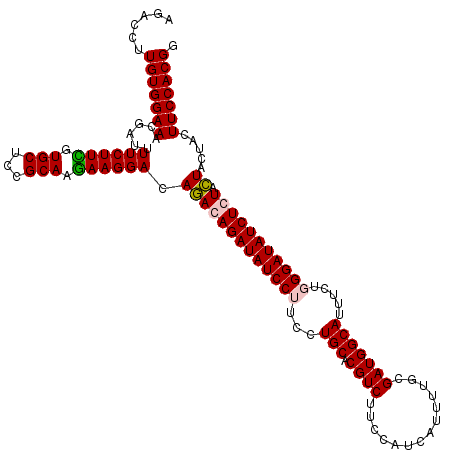
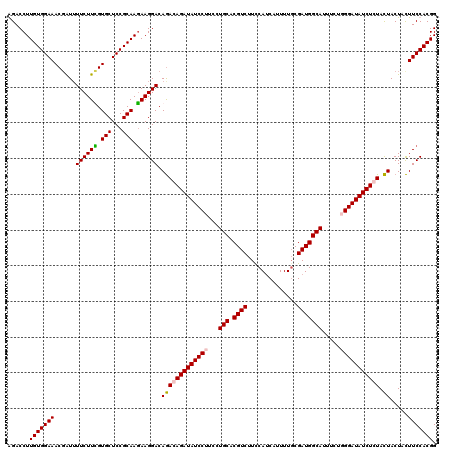
| Location | 5,640,294 – 5,640,414 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -37.69 |
| Consensus MFE | -34.62 |
| Energy contribution | -34.38 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5640294 120 - 27905053 CCGUGGAAGCAGUAGUAGAGAUAUCCAAAAAAUGCCAUCGCAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUUUUGCGGAGCACAAAGAAAAUGGUUUCCACAAGGUCU ..((((((((.............(((........((((((.....)))))).......((((((((((((......)))))))))))))))..............))))))))....... ( -38.23) >DroSec_CAF1 14121 120 - 1 CCGUGGAAGUAGUAGUACAGAUAUCCCAGAAAUGCCAUCGCAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACGAAGAAUAUCGUUUCCACAAGGUCC ..(((((((..(((.........(((........((((((.....)))))).......((((((((((((......)))))))))))))))..........)))..)))))))....... ( -40.01) >DroSim_CAF1 14825 120 - 1 CCGUGGAAGUAGUAGUAGAGAUAUCCCAGAAAUGCCAUCGCAAAAUGGUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACGAAGAAAAUCGUUUCCACAAGGUCU ..((((((...........(...(((.......(((((......))))).........((((((((((((......))))))))))))))).)((((......))))))))))....... ( -36.00) >DroYak_CAF1 15023 120 - 1 CCGUGGAAAAAGUAAUACAGAUAUCCGAAAAAUGCCAUCGAAAAGUGAUGAAAGACGUGCAGGAAGGAUAUCUGUUUGUCCUUCUUGCGGAGCACAAAGAAAACCGUUUCCACAAGGUCC ..(((((((..((..........(((.........(((((.....)))))........((((((((((((......)))))))))))))))...........))..)))))))....... ( -36.50) >consensus CCGUGGAAGUAGUAGUACAGAUAUCCCAAAAAUGCCAUCGCAAAAUGAUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUCCUUCUUGCGGAGCACAAAGAAAAUCGUUUCCACAAGGUCC ..(((((((..((..........(((........((((((.....)))))).......((((((((((((......)))))))))))))))...........))..)))))))....... (-34.62 = -34.38 + -0.25)



| Location | 5,640,334 – 5,640,445 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.60 |
| Mean single sequence MFE | -29.77 |
| Consensus MFE | -26.24 |
| Energy contribution | -26.87 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5640334 111 + 27905053 GACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUUUUGGAUAUCUCUACUACUGCUUCCACGGAUACGGUGGCGUUGCCUUUCCACAGUGCCUG--------- .......((((((((....(((.((((...((......)).)))))))......)))))))).....((((..((....))..))))(((((((........)))))))..--------- ( -26.60) >DroSec_CAF1 14161 111 + 1 GACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUCUGGGAUAUCUGUACUACUACUUCCACGGCUACGGUGGUGUUGCCUUUCCACAGUGCCUG--------- ...(((((((((((((...(((.((((...((......)).))))))).....))))))))))).))...........(((.((.((((..........)))).)))))..--------- ( -33.50) >DroSim_CAF1 14865 120 + 1 GACAGACAGAUAUCCUUCCUGCACGUCUUUCACCAUUUUGCGAUGGCAUUUCUGGGAUAUCUCUACUACUACUUCCACGGCUACGGUGGUGUUGCCUUUCCACAGUGCCUGCUAAACNNN .......(((((((((...(((.((((..............))))))).....)))))))))................(((.((.((((..........)))).)))))........... ( -27.34) >DroYak_CAF1 15063 111 + 1 GACAAACAGAUAUCCUUCCUGCACGUCUUUCAUCACUUUUCGAUGGCAUUUUUCGGAUAUCUGUAUUACUUUUUCCACGGAUACGGUGGAGUUGCAUUUCCACAGUGCCUG--------- .....((((((((((....(((.((((..............)))))))......))))))))))..............((.(((.((((((......)))))).)))))..--------- ( -31.64) >consensus GACAGACAGAUAUCCUUCCUGCACGUCUUCCAUCAUUUUGCGAUGGCAUUUCUGGGAUAUCUCUACUACUACUUCCACGGAUACGGUGGUGUUGCCUUUCCACAGUGCCUG_________ ...(((((((((((((...(((.((((..............))))))).....))))))))))).))...........((.(((.((((..........)))).)))))........... (-26.24 = -26.87 + 0.63)


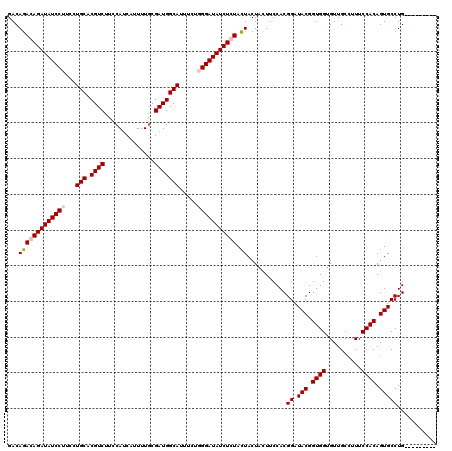
| Location | 5,640,334 – 5,640,445 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.60 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -25.17 |
| Energy contribution | -25.05 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5640334 111 - 27905053 ---------CAGGCACUGUGGAAAGGCAACGCCACCGUAUCCGUGGAAGCAGUAGUAGAGAUAUCCAAAAAAUGCCAUCGCAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUC ---------((((((((((.....(....).((((.......))))..))))).....((((((((........((((((.....))))))....(.....)...))))))))))))).. ( -32.40) >DroSec_CAF1 14161 111 - 1 ---------CAGGCACUGUGGAAAGGCAACACCACCGUAGCCGUGGAAGUAGUAGUACAGAUAUCCCAGAAAUGCCAUCGCAAAAUGAUGGAAGACGUGCAGGAAGGAUAUCUGUCUGUC ---------..(((((.((((...(....).)))).)).)))..........(((.((((((((((((....))((((((.....))))))..............))))))))))))).. ( -35.70) >DroSim_CAF1 14865 120 - 1 NNNGUUUAGCAGGCACUGUGGAAAGGCAACACCACCGUAGCCGUGGAAGUAGUAGUAGAGAUAUCCCAGAAAUGCCAUCGCAAAAUGGUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUC ........((((((.(..(((...((........))....)))..)............((((((((......((((.((.((......))...)).).)))....)))))))))))))). ( -29.40) >DroYak_CAF1 15063 111 - 1 ---------CAGGCACUGUGGAAAUGCAACUCCACCGUAUCCGUGGAAAAAGUAAUACAGAUAUCCGAAAAAUGCCAUCGAAAAGUGAUGAAAGACGUGCAGGAAGGAUAUCUGUUUGUC ---------..(((((..((((.(((.........))).))))..)..........((((((((((......((((.((....(....)....)).).)))....)))))))))).)))) ( -27.40) >consensus _________CAGGCACUGUGGAAAGGCAACACCACCGUAGCCGUGGAAGUAGUAGUACAGAUAUCCCAAAAAUGCCAUCGCAAAAUGAUGAAAGACGUGCAGGAAGGAUAUCUGUCUGUC .........(((((.(..(((...((........))....)))..)............((((((((......((((((((.....)))))........)))....))))))))))))).. (-25.17 = -25.05 + -0.12)

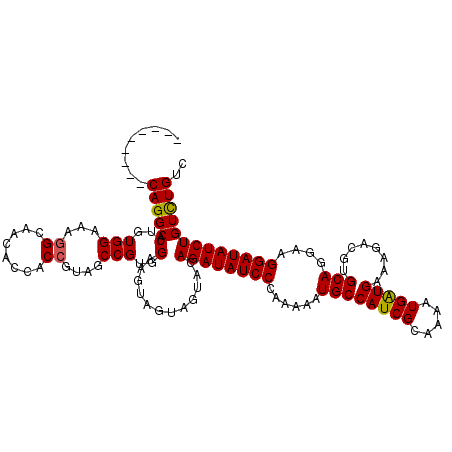

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:36 2006