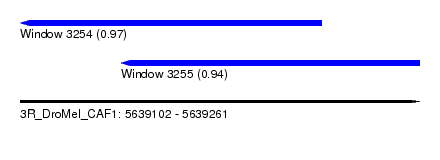
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,639,102 – 5,639,261 |
| Length | 159 |
| Max. P | 0.974936 |

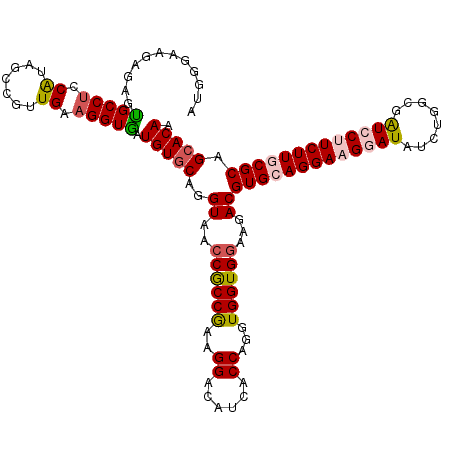
| Location | 5,639,102 – 5,639,222 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.42 |
| Mean single sequence MFE | -43.80 |
| Consensus MFE | -36.56 |
| Energy contribution | -37.32 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5639102 120 - 27905053 AUGGGAACAGAGUGCCUCCAUAGCCGUUGAAGGUAAUGUGCAGGUAUCCGCCAAAGGACAUCACCAGGUGGUGGAAGACGUGCAGGAAGGAUAUCUGGCGAUCCUUCUUGCGCAGCACAA .((....))...(((((.((.......)).))))).(((((..((.(((((((..((......))...)))))))..))(((((((((((((........))))))))))))).))))). ( -48.50) >DroSec_CAF1 12705 120 - 1 AUGGGAAGAGAGUGCCUCCAUAGCCGUUGAAGGUGAUGUGCAGGUAACCGCCGAAGGACAUCACCAGGUGGUGGAAGACGUGCAGGAAGGAUAUCUGGCGAUGCUUCUUGCGCAGCACAA ...........((((.(((((.(((......((((((((.(.(((....)))...).)))))))).))).)))))....((((((((((.((........)).)))))))))).)))).. ( -48.00) >DroSim_CAF1 13427 120 - 1 AUGGGAACAGAGUGCCUCCGUAGCCGUUGAAGGUGAUGUGCAGGUAUCCGCCGAAGGACAUCACCAGGUGGUGGAAGACGUGCAGGAAGGAUAUCUGGCGAUGCUUCUUGCGCAGCACAA .((....))..((((.((((..(((......((((((((.(.(((....)))...).)))))))).)))..))))....((((((((((.((........)).)))))))))).)))).. ( -47.50) >DroEre_CAF1 13798 120 - 1 AUGGGAAGAAAACGCCUCCAUAGCCGUUGAAGGUGAUGUUCACGUAGCCGCCAAAGGACAUCGCCAGGUGGUGAAAGACGUGCAGGACGGAUAUCUGGCGGUCCUUCUUUCGCAGCACAA .(((((((((..(((((((...(((......)))...((((((((.((((((...((......)).)))))).....)))))...)))))).....))))....))))))).))...... ( -36.20) >DroYak_CAF1 13589 120 - 1 AUGGGAAGAGAAUGCCGCCAUAGCCGCUGUAGGUGAUGUGCAUGUAACCACCGAAGGACAUGACCAGGUGGUGGAAGACGUGCAGGACGGAUAUCUGGCGAUCCUUCUUUCGCAGCACAA ..((((((.((.((((..(((.(((......))).)))(((((((..((((((..((......))...))))))...)))))))............)))).))))))))........... ( -38.80) >consensus AUGGGAAGAGAGUGCCUCCAUAGCCGUUGAAGGUGAUGUGCAGGUAACCGCCGAAGGACAUCACCAGGUGGUGGAAGACGUGCAGGAAGGAUAUCUGGCGAUCCUUCUUGCGCAGCACAA ............(((((.((.......)).))))).(((((..((..((((((..((......))...))))))...))(((((((((((((........))))))))))))).))))). (-36.56 = -37.32 + 0.76)

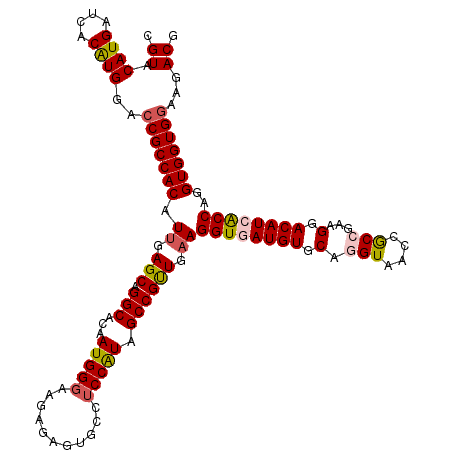
| Location | 5,639,142 – 5,639,261 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -42.01 |
| Consensus MFE | -35.78 |
| Energy contribution | -36.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.942202 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5639142 119 - 27905053 CGUACAUGAUCACAUGGACCGCCACAUUGAGCAGGCAUAAUGGGAACAGAGUGCCUCCAUAGCCGUUGAAGGUAAUGUGCAGGUAUCCGCCAAAGGACAUCACCAGGUGGUGGAAGACG (((.((((....))))..(((((((.(((.((((((((..((....))..)))))..(((.(((......))).)))))).(((.(((......))).)))..))))))))))...))) ( -41.80) >DroSec_CAF1 12745 119 - 1 CGUACAUGAUCACAUGGACCGCCACAUUGAGCAGGCACAAUGGGAAGAGAGUGCCUCCAUAGCCGUUGAAGGUGAUGUGCAGGUAACCGCCGAAGGACAUCACCAGGUGGUGGAAGACG (((.((((....))))..(((((((.((.(((.(((...(((((...........))))).)))))).))((((((((.(.(((....)))...).))))))))..)))))))...))) ( -42.70) >DroSim_CAF1 13467 119 - 1 CGUACAUGAUCACAUGGACCGCCACAUUGAGCAGGCACAAUGGGAACAGAGUGCCUCCGUAGCCGUUGAAGGUGAUGUGCAGGUAUCCGCCGAAGGACAUCACCAGGUGGUGGAAGACG (((.((((....))))..(((((((.....(.((((((..((....))..)))))).)............((((((((.(.(((....)))...).))))))))..)))))))...))) ( -47.40) >DroEre_CAF1 13838 119 - 1 UGUACAUGAUCACAUGGACCGCCACGUUGAGCAGGCAGGAUGGGAAGAAAACGCCUCCAUAGCCGUUGAAGGUGAUGUUCACGUAGCCGCCAAAGGACAUCGCCAGGUGGUGAAAGACG ....((((....)))).((((((...((.(((.(((...(((((...........))))).)))))).))((((((((((..............)))))))))).))))))........ ( -37.34) >DroYak_CAF1 13629 119 - 1 AGUACAUGAUCACGUGGACCGCCACGUUGAGCAGGCACGAUGGGAAGAGAAUGCCGCCAUAGCCGCUGUAGGUGAUGUGCAUGUAACCACCGAAGGACAUGACCAGGUGGUGGAAGACG .((((((.(((((((((....))))))..(((.(((...((((.............)))).))))))...))).))))))..((..((((((..((......))...))))))...)). ( -40.82) >consensus CGUACAUGAUCACAUGGACCGCCACAUUGAGCAGGCACAAUGGGAAGAGAGUGCCUCCAUAGCCGUUGAAGGUGAUGUGCAGGUAACCGCCGAAGGACAUCACCAGGUGGUGGAAGACG .((.((((....))))..(((((((.((.(((.(((...(((((...........))))).)))))).))((((((((.(.(((....)))...).))))))))..)))))))...)). (-35.78 = -36.38 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:31 2006