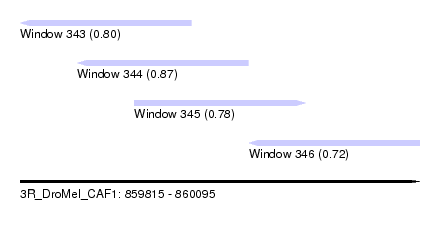
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 859,815 – 860,095 |
| Length | 280 |
| Max. P | 0.866192 |

| Location | 859,815 – 859,935 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -36.41 |
| Consensus MFE | -32.38 |
| Energy contribution | -33.88 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 859815 120 - 27905053 GGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAAAUAUGUAGGAAAUUAAGCUCACAGGCUCAACAACUUGCUC (((((((..((((......(((((((.....((((((....)))))).(((.((....))....)))..))))))).......(((..(........)..))).))))..)))..)))). ( -37.10) >DroSec_CAF1 34598 120 - 1 GGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGCGACCCUUCUCAAAAUAUGUAUGAAAUUAAGCUCACAGGCUCAACAACUUGCUC (((((((..((((.((...(((((((.....((((((....)))))).........((.(....).)).)))))))...........(((........))).))))))..)))..)))). ( -37.00) >DroSim_CAF1 33782 120 - 1 GGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAAAUAUGUAUGAAAUUAAGCCUACAGGCUCAACAACUUGCUC ((((..(((((((((...((...(((.....((((((....)))))).....))).)).(....)))).))))))....................((((....))))........)))). ( -37.80) >DroEre_CAF1 34772 119 - 1 GGCACUGGAGGACAAUCGCAGA-GGGGCAAAAGCCUCAAAGGGGGCUAACGACUUAUGGGGAAACGUGACCCUUCUCAAAAUAUGUGGGAAAUUAAGCUCACAGGCUCAACAACUUGGCC (((....(((.......(((((-((((((..((((((....))))))............(....).)).))))))).......((((((........)))))).)).......))).))) ( -33.74) >consensus GGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAAAUAUGUAGGAAAUUAAGCUCACAGGCUCAACAACUUGCUC (((((((..((((......(((((((.....((((((....))))))..........(.(....).)..))))))).......((((((........)))))).))))..)))..)))). (-32.38 = -33.88 + 1.50)

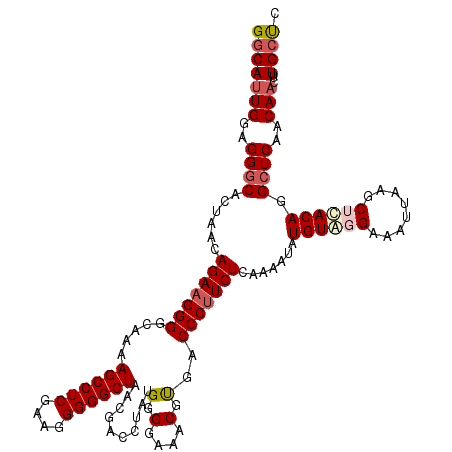
| Location | 859,855 – 859,975 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -32.56 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 859855 120 - 27905053 UGCUAUCAAAGCAAUCAAGUGAACAAAGGACAAGGCCUCUGGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAA ((((.....))))....((((..((.(((......))).)).))))(((((((((...((...(((.....((((((....)))))).....))).)).(....)))).))))))..... ( -37.50) >DroSec_CAF1 34638 120 - 1 UGCUAUCAAAGCAAUCAAGUGUACAAAGGACAAGGCCUCUGGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGCGACCCUUCUCAAA ..........((..((.(((((.((.(((......))).))))))).))..))......(((((((.....((((((....)))))).........((.(....).)).))))))).... ( -38.90) >DroSim_CAF1 33822 120 - 1 UGCUAUCAAAGCAAUCAAGUGAACAAAGGACAAGGCCUCUGGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAA ((((.....))))....((((..((.(((......))).)).))))(((((((((...((...(((.....((((((....)))))).....))).)).(....)))).))))))..... ( -37.50) >DroEre_CAF1 34812 119 - 1 UACUAUCAAAGCAAUCAAGUGAACAAAGGACAAGGCCUCUGGCACUGGAGGACAAUCGCAGA-GGGGCAAAAGCCUCAAAGGGGGCUAACGACUUAUGGGGAAACGUGACCCUUCUCAAA ..........((.....((((..((.(((......))).)).)))).((......))))(((-((((((..((((((....))))))............(....).)).))))))).... ( -30.90) >consensus UGCUAUCAAAGCAAUCAAGUGAACAAAGGACAAGGCCUCUGGCAUUGGAGGGCACUAACAGAAGGGGCAAAAGCCUCGAAGGGGGCUAACGACCUAUGGGGAAACGUGACCCUUCUCAAA ((((.....))))....................(.(((((......))))).)......(((((((.....((((((....))))))..........(.(....).)..))))))).... (-32.56 = -32.88 + 0.31)

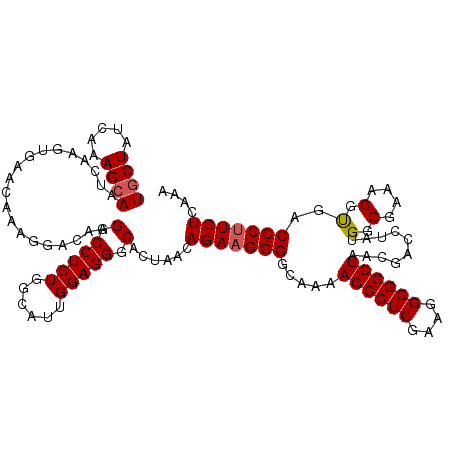
| Location | 859,895 – 860,015 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.96 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -37.97 |
| Energy contribution | -38.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 859895 120 + 27905053 UUCGAGGCUUUUGCCCCUUCUGUUAGUGCCCUCCAAUGCCAGAGGCCUUGUCCUUUGUUCACUUGAUUGCUUUGAUAGCAUGGAGGGUACGGUAUCGUGGUGAGGGGUUCGAGGUUCGAG .(((((.(((..((((((((..(..((((((((((....((((((......))))))..........((((.....))))))))))))))......)..).)))))))..))).))))). ( -50.20) >DroSec_CAF1 34678 117 + 1 UUCGAGGCUUUUGCCCCUUCUGUUAGUGCCCUCCAAUGCCAGAGGCCUUGUCCUUUGUACACUUGAUUGCUUUGAUAGCAUCAAGGGUACGGAAUCGUGGUGAGGGGUUCGAGGUUC--- ...(((.(((..((((((((..(..((((((........((((((......)))))).....(((((.(((.....))))))))))))))......)..).)))))))..))).)))--- ( -38.90) >DroSim_CAF1 33862 117 + 1 UUCGAGGCUUUUGCCCCUUCUGUUAGUGCCCUCCAAUGCCAGAGGCCUUGUCCUUUGUUCACUUGAUUGCUUUGAUAGCAUCGAGGGUACGGAAUCGUGUUGAGGGGUACGAGGUUC--- ...(((.(((.(((((((((.(((.((((((((......((((((......)))))).......(((.(((.....))))))))))))))..))).)....)))))))).))).)))--- ( -41.60) >consensus UUCGAGGCUUUUGCCCCUUCUGUUAGUGCCCUCCAAUGCCAGAGGCCUUGUCCUUUGUUCACUUGAUUGCUUUGAUAGCAUCGAGGGUACGGAAUCGUGGUGAGGGGUUCGAGGUUC___ ...(((.(((..(((((((.((...(((((((((((((.((((((......))))))..)).)))..((((.....))))..)))))))).....))....)))))))..))).)))... (-37.97 = -38.30 + 0.33)

| Location | 859,975 – 860,095 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.44 |
| Mean single sequence MFE | -37.33 |
| Consensus MFE | -30.53 |
| Energy contribution | -32.03 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 0.97 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 859975 120 - 27905053 ACCGCAGUAGUCCUCCUGUGCACCGAAGAGGAAGUAAGCCGGACUACCGUGGAGGGUACCUCGAACCUCGAACCUCGAACCUCGAACCUCGAACCCCUCACCACGAUACCGUACCCUCCA ...((.(((((((..((.(((.((.....))..)))))..))))))).))(((((((((.((((...(((.....)))...))))...(((............)))....))))))))). ( -38.40) >DroSec_CAF1 34758 106 - 1 ACCGCAGUAGUCCUCCUGUGCACCGAAGGGGAAGUAAGCCGGACUACCGUGGAGGGUACCUCGAACCUC--------------GAACCUCGAACCCCUCACCACGAUUCCGUACCCUUGA .((((.(((((((..((.(((.((....))...)))))..))))))).))))(((((((.(((.....)--------------))...(((............)))....)))))))... ( -35.60) >DroSim_CAF1 33942 106 - 1 ACCGCAGUAGUCCUCCUGUGCACCGAAGGGGAAGUAAGCCGGACUACCGUGGAGGGUACCUCGAACCUC--------------GAACCUCGUACCCCUCAACACGAUUCCGUACCCUCGA ..(((.(((((((..((.(((.((....))...)))))..))))))).)))((((((((.(((.....)--------------))...((((..........))))....)))))))).. ( -38.00) >consensus ACCGCAGUAGUCCUCCUGUGCACCGAAGGGGAAGUAAGCCGGACUACCGUGGAGGGUACCUCGAACCUC______________GAACCUCGAACCCCUCACCACGAUUCCGUACCCUCGA .((((.(((((((..((.(((.((....))...)))))..))))))).))))(((((((.((((...(((((........)))))...)))).....((.....))....)))))))... (-30.53 = -32.03 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:08 2006