
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,547,924 – 5,548,077 |
| Length | 153 |
| Max. P | 0.850260 |

| Location | 5,547,924 – 5,548,037 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 83.15 |
| Mean single sequence MFE | -41.66 |
| Consensus MFE | -27.96 |
| Energy contribution | -29.00 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
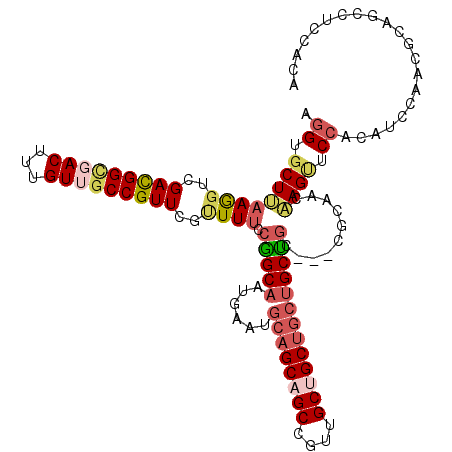
>3R_DroMel_CAF1 5547924 113 - 27905053 UCCGGCAAUAAAUGCAGCAGCCGUUGCUGCUGCUGCUGC---CGCAACAAGUUCCACAUCUAACACAGCCUCCACAUCCAAGGAAGAGGGCAAGGAGGACGACUGGCAGCAGUGUC ...((((......((((((((....))))))))((((((---(.......(((........)))...((((((...(((........)))...))))).)....))))))).)))) ( -41.20) >DroSec_CAF1 7007 113 - 1 UCCGGCAAUAAAUGCAGCAGCCGUUGCUGCUGCUGCCGC---CGCAACAAGUUCCACAUCCAACGCACCCUCCACAUCCAAGGAAGAGGGCAAGGAGGAUGACUGGCAGCAAUGUC ...((((......((((((((....))))))))(((.((---(((.....))....(((((..(...(((((....((....)).)))))...)..)))))...))).))).)))) ( -41.70) >DroEre_CAF1 6213 116 - 1 UCCGGCAAUGAAUGCAGCAGCAGUUGCUGCUGCUGCUGCCGCCGCAACAAGUUCCUCAUCCAACGCAGCCUCCACAUCCAAGGAAGAGGGGAAGGAGGACGAUUGGCAGCAGUGUC ...((((......((((((((....))))))))((((((((.(((..(...((((((.(((....................))).))))))...)..).))..)))))))).)))) ( -43.65) >DroYak_CAF1 6233 113 - 1 UCCAGCAAUGAAUGCAGCAGCCGUUGCCGCUGCUGCUGC---CGCAUCAAGUUCCUCAUCCAACACAGCCUCCACAUCCAAGGAAGAGGGAAAGGAGGACGACUGGCAGCAGUGUC ....((((((...((....))))))))(((((((((((.---(((.((...((((((.(((....................))).))))))...)).).)).).)))))))))).. ( -37.85) >DroAna_CAF1 6932 116 - 1 CCCGGCAAUGAAUCAAGCAGCACCUGCAGCCGUGGCUUCCAACUCGGCUAGCACCAGCUCGAGUGCUGCUUCCACAUCGAAGGAGGAGGGCAAGGAAGAUGACUGGCAGCAGUGCC ...((((.((......((((...)))).(((((.((((((.((((((((......))).)))))(((.(((((........)))))..)))..))))).).)).)))..)).)))) ( -43.90) >consensus UCCGGCAAUGAAUGCAGCAGCCGUUGCUGCUGCUGCUGC___CGCAACAAGUUCCACAUCCAACGCAGCCUCCACAUCCAAGGAAGAGGGCAAGGAGGACGACUGGCAGCAGUGUC ...((((......((((((((....)))))))).(((((...........(((........)))....(((((...(((........)))...))))).......)))))..)))) (-27.96 = -29.00 + 1.04)



| Location | 5,547,964 – 5,548,077 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -43.66 |
| Consensus MFE | -28.84 |
| Energy contribution | -30.24 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5547964 113 + 27905053 UGUGGAGGCUGUGUUAGAUGUGGAACUUGUUGCG---GCAGCAGCAGCAGCAACGGCUGCUGCAUUUAUUGCCGGAAAACGAACGGCAACAAAGUCGCCGUCGACCUUAAGCACCU ...((..(((..(((........)))..((((((---((.((.((((((((....)))))))).(((.((((((.........)))))).))))).)))).))))....))).)). ( -42.30) >DroSec_CAF1 7047 113 + 1 UGUGGAGGGUGCGUUGGAUGUGGAACUUGUUGCG---GCGGCAGCAGCAGCAACGGCUGCUGCAUUUAUUGCCGGAAAUCGAACGGCCACAAAGUCUCCGUCGACCUUAAGAACCU .((..(((((.((.(((((((((....((((.((---((((((((((((((....))))))))......)))))....))))))).))))).....)))).)))))))....)).. ( -41.40) >DroEre_CAF1 6253 116 + 1 UGUGGAGGCUGCGUUGGAUGAGGAACUUGUUGCGGCGGCAGCAGCAGCAGCAACUGCUGCUGCAUUCAUUGCCGGAAAACGAACGGCAACAAAGUCGCCGUCGACCUUAAGCACCU .....(((.((((((........)))..(((((((((((.((((((((((...)))))))))).....((((((.........))))))....))))))).)))).....)))))) ( -51.70) >DroYak_CAF1 6273 113 + 1 UGUGGAGGCUGUGUUGGAUGAGGAACUUGAUGCG---GCAGCAGCAGCGGCAACGGCUGCUGCAUUCAUUGCUGGAAAACGAACGGCAACAAAGUCACCGUCGACCUUUAGCACCU .(((((.(((((((..(.........)..)))))---)).(((((((((....).)))))))).)))))(((((((...(((.(((..((...))..))))))...)))))))... ( -41.60) >DroAna_CAF1 6972 116 + 1 UGUGGAAGCAGCACUCGAGCUGGUGCUAGCCGAGUUGGAAGCCACGGCUGCAGGUGCUGCUUGAUUCAUUGCCGGGAAGCGAACGGCUACAAAGUCACCAUCUACUUUCAGCACUU .(((((((((((((..((((((..(((..((.....)).)))..))))).)..))))))))(((((....((((.........)))).....)))))...)))))........... ( -41.30) >consensus UGUGGAGGCUGCGUUGGAUGUGGAACUUGUUGCG___GCAGCAGCAGCAGCAACGGCUGCUGCAUUCAUUGCCGGAAAACGAACGGCAACAAAGUCACCGUCGACCUUAAGCACCU .....(((.(((....((((.(..(((((((((....))))).((((((((....)))))))).....((((((.........))))))..))))..)))))........)))))) (-28.84 = -30.24 + 1.40)



| Location | 5,547,964 – 5,548,077 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -26.90 |
| Energy contribution | -28.62 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5547964 113 - 27905053 AGGUGCUUAAGGUCGACGGCGACUUUGUUGCCGUUCGUUUUCCGGCAAUAAAUGCAGCAGCCGUUGCUGCUGCUGCUGC---CGCAACAAGUUCCACAUCUAACACAGCCUCCACA .((.((((..((.((((((((((...))))))).)))....))((((......((((((((....))))))))...)))---).....)))).))..................... ( -41.00) >DroSec_CAF1 7047 113 - 1 AGGUUCUUAAGGUCGACGGAGACUUUGUGGCCGUUCGAUUUCCGGCAAUAAAUGCAGCAGCCGUUGCUGCUGCUGCCGC---CGCAACAAGUUCCACAUCCAACGCACCCUCCACA ..........(((((..((((((((((((((.....(.....)((((......((((((((....))))))))))))))---))))..))))).....)))..)).)))....... ( -37.10) >DroEre_CAF1 6253 116 - 1 AGGUGCUUAAGGUCGACGGCGACUUUGUUGCCGUUCGUUUUCCGGCAAUGAAUGCAGCAGCAGUUGCUGCUGCUGCUGCCGCCGCAACAAGUUCCUCAUCCAACGCAGCCUCCACA ((((((.....((.(.(((((..((..((((((.........))))))..)).(((((((((((....))))))))))))))))).))..(((........)))))).)))..... ( -47.60) >DroYak_CAF1 6273 113 - 1 AGGUGCUAAAGGUCGACGGUGACUUUGUUGCCGUUCGUUUUCCAGCAAUGAAUGCAGCAGCCGUUGCCGCUGCUGCUGC---CGCAUCAAGUUCCUCAUCCAACACAGCCUCCACA .(((((....((.((((((..((...))..))).)))....))..........((((((((.((....)).))))))))---.)))))............................ ( -35.80) >DroAna_CAF1 6972 116 - 1 AAGUGCUGAAAGUAGAUGGUGACUUUGUAGCCGUUCGCUUCCCGGCAAUGAAUCAAGCAGCACCUGCAGCCGUGGCUUCCAACUCGGCUAGCACCAGCUCGAGUGCUGCUUCCACA ...(((((.(((..((((((.((...)).))))))..)))..))))).......((((((((((((((((((.(........).))))).))).......).)))))))))..... ( -40.91) >consensus AGGUGCUUAAGGUCGACGGCGACUUUGUUGCCGUUCGUUUUCCGGCAAUGAAUGCAGCAGCCGUUGCUGCUGCUGCUGC___CGCAACAAGUUCCACAUCCAACGCAGCCUCCACA .((.((((((((..(((((((((...)))))))))..)))).(((((......((((((((....)))))))))))))..........)))).))..................... (-26.90 = -28.62 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:24:01 2006