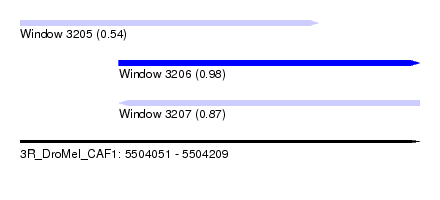
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,504,051 – 5,504,209 |
| Length | 158 |
| Max. P | 0.984794 |

| Location | 5,504,051 – 5,504,169 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.09 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -29.46 |
| Energy contribution | -29.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
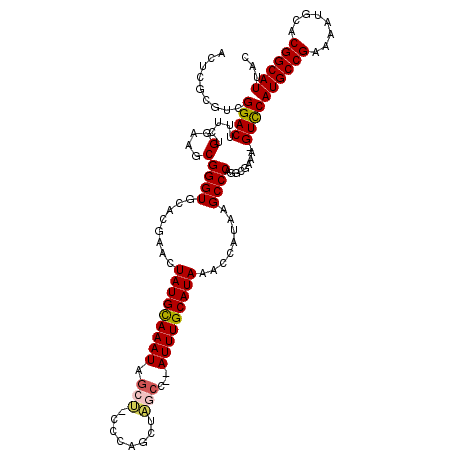
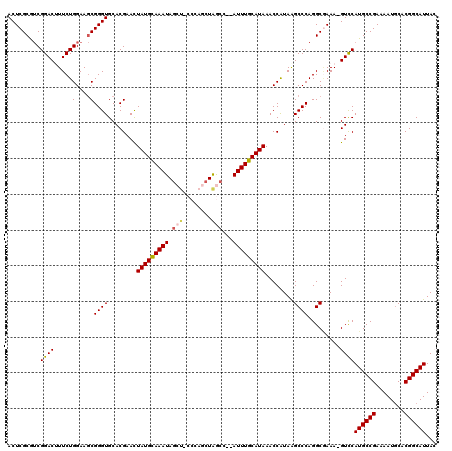
>3R_DroMel_CAF1 5504051 118 + 27905053 CGGAGGGUCCAGUGUUUGAGGUUAUGUU-UUCGGCUGGCGGUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUUGCCUGGGCUUAUGGUUUAUGCAAAUCUAGCUAGCUGGGAAGCUAUUUA .((.....))................((-..((((((((((.(((((...))))).)).((((((((-(...((....))....))))))))).......))))))))..))........ ( -37.20) >DroSec_CAF1 3743 116 + 1 CGGAGGGUCCAGUGUUUGAGGUUAUGUU-UUCGGCUGGCGGUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCCUACGGUUUAUGCAAAU--GGCUAGCUGGGCAGCUAUUUG ((..((((((((.((..(((..((((((-....(((((((....))))).))......))))))..)-))..)))))))))).)).......(((((--((((.((...))))))))))) ( -42.20) >DroSim_CAF1 4204 116 + 1 CGGAGGGUCCAGUGUUUGAGGUUAUGUU-UUCGGCUGGCGGUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCCUACGGUUUAUGCAAAU--GGCUAGCUGGGCAGCUAUUUG ((..((((((((.((..(((..((((((-....(((((((....))))).))......))))))..)-))..)))))))))).)).......(((((--((((.((...))))))))))) ( -42.20) >DroEre_CAF1 3717 114 + 1 CGGAUGGUCCAGUCUUUGAGGUUAUGUU-UUCGGCUGGCUCUAAUGCCGUGGAUUUUCGGCAUGAACCUUUGGCCUGGGCUUAUGGUUUAUGCAAAU--GGUCAGUUC---UGCUAUUUG ((((.(((((((((...((((......)-)))))))(((......)))..)))))))))(((((((((...(((....)))...)))))))))((((--((.((....---)))))))). ( -37.90) >DroYak_CAF1 3876 114 + 1 CGGAGGGUCCAGUCUUUGAGGUUAUGUUCUUCGGCUGGCGCUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCUUAUGGUUUAUGCAAAU--GGUCAGCUC---GUUUAUUUG ..((((((((.(((.((((((.......))))))..)))....((((((........))))))))))-))))....(((((.((.((((....))))--.)).)))))---......... ( -36.20) >consensus CGGAGGGUCCAGUGUUUGAGGUUAUGUU_UUCGGCUGGCGGUAAUGCCGUGCAUUUUCGGCAUGGAC_UUUCGCCUGGGCUUAUGGUUUAUGCAAAU__GGCUAGCUGGG_AGCUAUUUG .((.....))........((((...(((.((((((((((....((((((........)))))).........((.((((((...)))))).)).......)))))))))).))).)))). (-29.46 = -29.90 + 0.44)

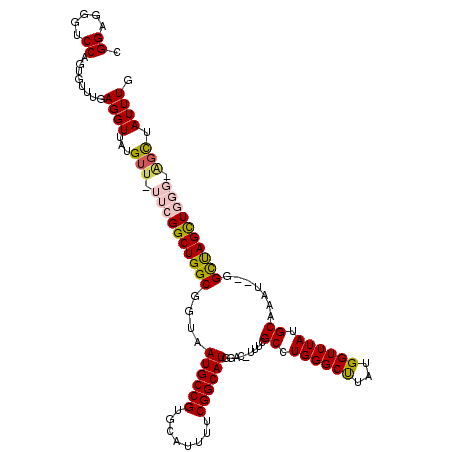
| Location | 5,504,090 – 5,504,209 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -33.72 |
| Energy contribution | -33.44 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.984794 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5504090 119 + 27905053 GUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUUGCCUGGGCUUAUGGUUUAUGCAAAUCUAGCUAGCUGGGAAGCUAUUUACAUAGUUCGUGCACCCGCUUCCAGAAAGUCCGACGCGAGU ...((((((........))))))((((-((((((...((((...(((((....))))).)))).))..((((((....(((.......))).....)))))).)))))))).((....)) ( -36.90) >DroSec_CAF1 3782 117 + 1 GUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCCUACGGUUUAUGCAAAU--GGCUAGCUGGGCAGCUAUUUGCAUAGUUCGUGCACCCGCUUCCAGAAUGUCCGACGCGAGU ...((((((........))))))((((-.(((...((((..((((..((((((((((--((((.((...))))))))))))))))..))))..))))......))).)))).((....)) ( -48.20) >DroSim_CAF1 4243 117 + 1 GUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCCUACGGUUUAUGCAAAU--GGCUAGCUGGGCAGCUAUUUGCAUAGUUCGUGCACCCGCUUCCAGAAAGUCCGACGCGAGU ...((((((........))))))((((-((((...((((..((((..((((((((((--((((.((...))))))))))))))))..))))..))))......)))))))).((....)) ( -52.30) >DroEre_CAF1 3756 115 + 1 CUAAUGCCGUGGAUUUUCGGCAUGAACCUUUGGCCUGGGCUUAUGGUUUAUGCAAAU--GGUCAGUUC---UGCUAUUUGCAUAAUUCGUGCACCCGGUUCCAGAAAGUCCGUUGCGAGU ....(((..(((((((((((......))..(((.(((((..((((..((((((((((--((.((....---))))))))))))))..))))..)))))..))))))))))))..)))... ( -45.90) >DroYak_CAF1 3916 114 + 1 CUAAUGCCGUGCAUUUUCGGCAUGGAC-UUUCGCCUGGGCUUAUGGUUUAUGCAAAU--GGUCAGCUC---GUUUAUUUGCAUAGUUCGUGCACCCGCUUCCAGAAUGUCCGACGCGAGU ...((((((........))))))((((-.(((...((((..((((..((((((((((--((.(.....---).))))))))))))..))))..))))......))).)))).((....)) ( -38.10) >consensus GUAAUGCCGUGCAUUUUCGGCAUGGAC_UUUCGCCUGGGCUUAUGGUUUAUGCAAAU__GGCUAGCUGGG_AGCUAUUUGCAUAGUUCGUGCACCCGCUUCCAGAAAGUCCGACGCGAGU ...((((((........))))))((((.(((....((((..((((..((((((((((..((((........))))))))))))))..))))..))))......))).))))......... (-33.72 = -33.44 + -0.28)


| Location | 5,504,090 – 5,504,209 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.80 |
| Mean single sequence MFE | -37.29 |
| Consensus MFE | -27.57 |
| Energy contribution | -27.81 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871790 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5504090 119 - 27905053 ACUCGCGUCGGACUUUCUGGAAGCGGGUGCACGAACUAUGUAAAUAGCUUCCCAGCUAGCUAGAUUUGCAUAAACCAUAAGCCCAGGCAAAA-GUCCAUGCCGAAAAUGCACGGCAUUAC .........(((((((((((..((.(((........(((((((((((((........)))))..)))))))).)))....)))))))...))-))))((((((........))))))... ( -34.80) >DroSec_CAF1 3782 117 - 1 ACUCGCGUCGGACAUUCUGGAAGCGGGUGCACGAACUAUGCAAAUAGCUGCCCAGCUAGCC--AUUUGCAUAAACCGUAGGCCCAGGCGAAA-GUCCAUGCCGAAAAUGCACGGCAUUAC ....((.((((.....))))..))((((..(((...(((((((((.(((((...)).))).--)))))))))...)))..)))).(((....-))).((((((........))))))... ( -40.90) >DroSim_CAF1 4243 117 - 1 ACUCGCGUCGGACUUUCUGGAAGCGGGUGCACGAACUAUGCAAAUAGCUGCCCAGCUAGCC--AUUUGCAUAAACCGUAGGCCCAGGCGAAA-GUCCAUGCCGAAAAUGCACGGCAUUAC .........((((((((.(....)((((..(((...(((((((((.(((((...)).))).--)))))))))...)))..))))....))))-))))((((((........))))))... ( -44.00) >DroEre_CAF1 3756 115 - 1 ACUCGCAACGGACUUUCUGGAACCGGGUGCACGAAUUAUGCAAAUAGCA---GAACUGACC--AUUUGCAUAAACCAUAAGCCCAGGCCAAAGGUUCAUGCCGAAAAUCCACGGCAUUAG .........(((((((.(((..((((((.......((((((((((.(((---....)).).--)))))))))).......)))).))))))))))))((((((........))))))... ( -35.84) >DroYak_CAF1 3916 114 - 1 ACUCGCGUCGGACAUUCUGGAAGCGGGUGCACGAACUAUGCAAAUAAAC---GAGCUGACC--AUUUGCAUAAACCAUAAGCCCAGGCGAAA-GUCCAUGCCGAAAAUGCACGGCAUUAG ....((.((((.....))))..))((((........(((((((((....---.........--)))))))))........)))).(((....-))).((((((........))))))... ( -30.91) >consensus ACUCGCGUCGGACUUUCUGGAAGCGGGUGCACGAACUAUGCAAAUAGCU_CCCAGCUAGCC__AUUUGCAUAAACCAUAAGCCCAGGCGAAA_GUCCAUGCCGAAAAUGCACGGCAUUAC .........((((.....(....)((((........(((((((((.(((........)))...)))))))))........)))).........))))((((((........))))))... (-27.57 = -27.81 + 0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:42 2006