
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,462,765 – 5,462,874 |
| Length | 109 |
| Max. P | 0.927672 |

| Location | 5,462,765 – 5,462,874 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -30.85 |
| Energy contribution | -32.60 |
| Covariance contribution | 1.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.605530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5462765 109 + 27905053 AUGUGGAUGUGGAGGAGGCGGAUGCGGAUGCGGAGGAGGUGGCAUUCGUACUGGCCCCUGGCGCAGCAGAAUUGCUCCCGCUACUGGUACUGCUAUCACGUCCGCCCGA ..((((((((((((.((.(((.(((((((((.(......).))))))))))))(((..(((((.(((......)))..)))))..))).)).)).)))))))))).... ( -45.60) >DroSec_CAF1 2712 103 + 1 AUGUGGAUGUGGAGGAGGCGGA------UGCGGAGGAGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGUUGCUGCUAUCACGUCCGCCUGA ..(((((((((((((((.(((.------((((((.(......).))))))))).)))))((.(((((((....((....))..))))))).))..)))))))))).... ( -48.00) >DroEre_CAF1 1968 97 + 1 AUGUGGAUGUGGAGGAGGCG------------GAGGAUGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGUUGCUGCUAUCACGUCCGCCCGA ..(((((((((((((((.((------------(.(((((...)))))...))).)))))((.(((((((....((....))..))))))).))..)))))))))).... ( -44.80) >DroYak_CAF1 1413 97 + 1 AUGUGGAUGUGGAGGAGGCG------------GAGGUGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGGUGCUGCUAUCACGUCCGCCGGA ................((((------------((.(((((((((...((((((((....)))(((((.((.....))..))))).)))))))))))))).))))))... ( -46.30) >consensus AUGUGGAUGUGGAGGAGGCG____________GAGGAGGUGGCAUUCGUACUGGCUCCUGCCGCAGCAGAAUUGCUCCCGCUGCUGGUGCUGCUAUCACGUCCGCCCGA ..(((((((((((((((.(((.............................))).)))))((.(((((((....((....))..))))))).))..)))))))))).... (-30.85 = -32.60 + 1.75)

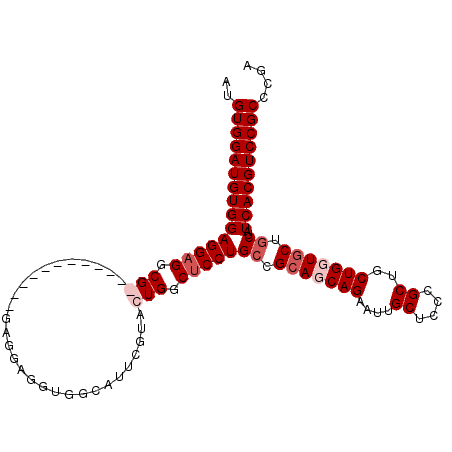
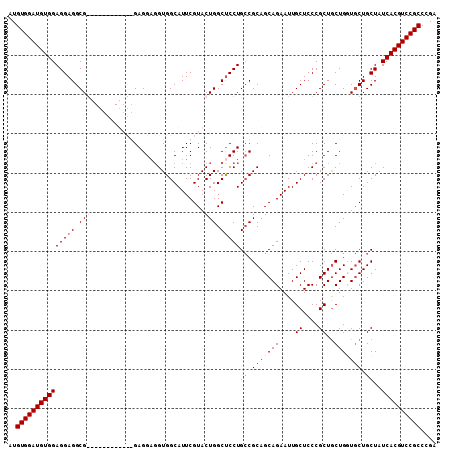
| Location | 5,462,765 – 5,462,874 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 89.05 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -30.19 |
| Energy contribution | -30.62 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5462765 109 - 27905053 UCGGGCGGACGUGAUAGCAGUACCAGUAGCGGGAGCAAUUCUGCUGCGCCAGGGGCCAGUACGAAUGCCACCUCCUCCGCAUCCGCAUCCGCCUCCUCCACAUCCACAU ..(((((((.(((((.((.......(((((((((....)))))))))...((((((..........))).))).....)))).))).)))))))............... ( -38.40) >DroSec_CAF1 2712 103 - 1 UCAGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCUCCUCCGCA------UCCGCCUCCUCCACAUCCACAU ..(((((((.((....)).((....(((((((((....)))))))))((.(((((...(((....)))...))))))))).------)))))))............... ( -40.90) >DroEre_CAF1 1968 97 - 1 UCGGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACAUCCUC------------CGCCUCCUCCACAUCCACAU ..(((((((.(.(((.(..(((...(((((((((....)))))))))(((....)))........)))..)))))))------------)))))............... ( -37.70) >DroYak_CAF1 1413 97 - 1 UCCGGCGGACGUGAUAGCAGCACCAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCACCUC------------CGCCUCCUCCACAUCCACAU ...((((((.(((...(((......(((((((((....)))))))))(((....)))........)))...))).))------------))))................ ( -39.80) >consensus UCGGGCGGACGUGAUAGCAGCAACAGCAGCGGGAGCAAUUCUGCUGCGGCAGGAGCCAGUACGAAUGCCACCUCCUC____________CGCCUCCUCCACAUCCACAU ...((.(((.(((......((....(((((((((....))))))))).))(((((...(((....)))...))))).............))).))).)).......... (-30.19 = -30.62 + 0.44)

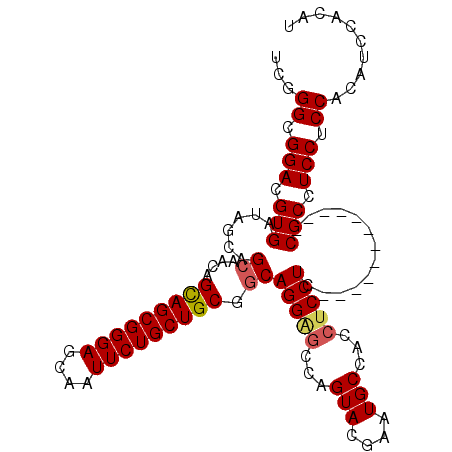
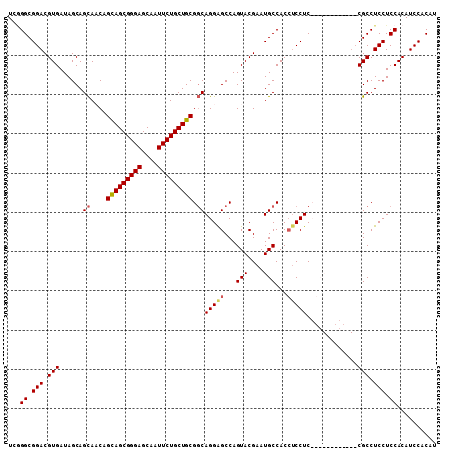
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:17 2006