| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
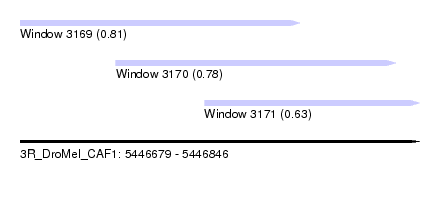
| Location | 5,446,679 – 5,446,846 |
| Length | 167 |
| Max. P | 0.814279 |

| Location | 5,446,679 – 5,446,796 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.37 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -20.83 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5446679 117 + 27905053 CACAACACAACACGAGACGAGACGAGACCGAAAGGGAAAAAAACACUUUAAAGCUGC---UGUCGAGUGUUAAUCAUGGUGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGC .......((((................((....)).................((.((---((..((((((((((..((((...........))))))))))))))..)))).)).)))). ( -29.30) >DroEre_CAF1 33840 99 + 1 CACAACA-----CGAG----------ACCAAGAGG--AA-AAACACUUUACAGCUGC---UGCCGAGUGUUAAUCAUGGGGUAAUUUAUUAGUCAGUUGACACUUUUUGGUCUCAGUUGC ..((((.-----.(((----------(((((((((--..-.(((((((..(((...)---))..)))))))....................(((....))).)))))))))))).)))). ( -31.50) >DroYak_CAF1 34539 109 + 1 CACAACA-----CGAGAC----UGAGACCAAGAGG--AAAAAACACUUUACAGCUGCUGCUGCCAAGUGUUAAUCAUGGGGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGC .......-----...(((----((.(.((((((((--....(((((((..((((....))))..)))))))..(((..((.(((....))).))...)))..)))))))).).))))).. ( -33.60) >consensus CACAACA_____CGAGAC_____GAGACCAAGAGG__AAAAAACACUUUACAGCUGC___UGCCGAGUGUUAAUCAUGGGGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGC ...........................((....))...............(((((((....(((((((((((((..(((.............)))))))))))))...))).))))))). (-20.83 = -20.62 + -0.21)



| Location | 5,446,719 – 5,446,836 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -30.40 |
| Energy contribution | -30.19 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5446719 117 + 27905053 AAACACUUUAAAGCUGC---UGUCGAGUGUUAAUCAUGGUGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAU ...((((....((((((---.(((((((((((((..((((...........))))))))))))))...))).))))))...........((((((....))))))......))))..... ( -34.80) >DroEre_CAF1 33862 117 + 1 AAACACUUUACAGCUGC---UGCCGAGUGUUAAUCAUGGGGUAAUUUAUUAGUCAGUUGACACUUUUUGGUCUCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAU ...(((((..((((((.---.(((((((((((((....((.(((....))).)).))))))))))...)))..))))))..))......((((((....)))))).......)))..... ( -33.10) >DroYak_CAF1 34568 120 + 1 AAACACUUUACAGCUGCUGCUGCCAAGUGUUAAUCAUGGGGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAU ...((((...((((.(((((.(((((((((((((....((.(((....))).)).)))))))))...)))).))))).)).........((((((....))))))))....))))..... ( -41.00) >consensus AAACACUUUACAGCUGC___UGCCGAGUGUUAAUCAUGGGGUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAU ...(((((..(((((((....(((((((((((((..(((.............)))))))))))))...))).)))))))..))......((((((....)))))).......)))..... (-30.40 = -30.19 + -0.21)


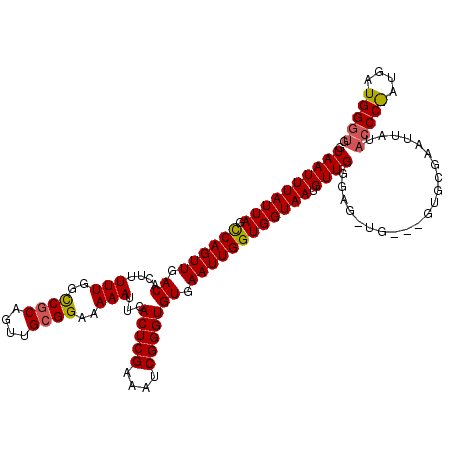
| Location | 5,446,756 – 5,446,846 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -22.10 |
| Energy contribution | -22.80 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631236 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5446756 90 + 27905053 GUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAUGUUGGGA------------------------------GUG .((((((((((.((((((.((....(((..((((....))))..)))...(((((....))))))).)))))))))))).))))...------------------------------... ( -25.00) >DroEre_CAF1 33899 120 + 1 GUAAUUUAUUAGUCAGUUGACACUUUUUGGUCUCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAUGUUGGCAGUUGAGUGUGCGAAUUAUACCCAAUGAUGGGUG ................(((((((((....(((...(((((.(...((((((((((....)))..)))))))..).)))))...)))....)))))).)))....((((((....)))))) ( -32.00) >DroYak_CAF1 34608 120 + 1 GUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAUGUUGCGAGCUGGAAGUGCGAAUUAUACCCGAUGAUGGGUG (((((((((((.((((((.((....(((..((((....))))..)))...(((((....))))))).)))))))))))).)))))..((.......))......(((((......))))) ( -35.40) >consensus GUAAUUUAUUAGCCAGUUGACACUUUUUGGCCGCAGUUGCGGAAAAAUUCACUCGAAAUCGGGUGUGAAUUGGUGGUAAUGUUGGGAG_UG___GUGCGAAUUAUACCC_AUGAUGGGUG .((((((((((.((((((.((....(((..((((....))))..)))...(((((....))))))).)))))))))))).)))).....................(((((....))))). (-22.10 = -22.80 + 0.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:05 2006