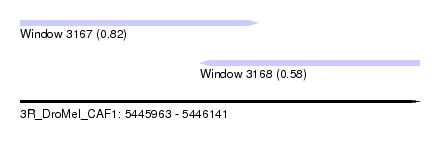
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,445,963 – 5,446,141 |
| Length | 178 |
| Max. P | 0.816691 |

| Location | 5,445,963 – 5,446,069 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.86 |
| Mean single sequence MFE | -24.00 |
| Consensus MFE | -19.63 |
| Energy contribution | -18.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5445963 106 + 27905053 CACUUGGCCGUAAAUUACCAUAAAUUCCAUUCCGCAGAACAGUUUGAUAUUUAUAAUUUUAAAUCACGAGCACGACUGUCUGCAACU--------------UUAUAGUUGGCCAUAAAAA ....(((((........................(((((.(((((((..(((((......)))))......)).))))))))))((((--------------....)))))))))...... ( -23.40) >DroEre_CAF1 33114 120 + 1 CACUUGGCCGUAAAUUACCAUAAAUUCCAUUCCGCGGAACAGUUUAAUAUUUAUAAUUCUAAAUCACGAGCACGACUGUCUGCAGCUUUGUAUGUACUCGUACAUAGUUGGCCAUAAAAA ....(((((........................(((((.((((.....(((((......)))))..((....)))))))))))((((.((((((....)))))).)))))))))...... ( -27.90) >DroYak_CAF1 33771 106 + 1 CACUUGGCCGUAAAUUACCAUAAAUUCCAUUCCGCAGAACAGUUUAAUAUUUAUAAUUCUAAAUCACGAGCACGACUGUCUGCAGCU--------------AUAUGGUUGGCCAUAAAAA ....((((((......(((((((((...)))..(((((.((((.....(((((......)))))..((....)))))))))))....--------------.))))))))))))...... ( -20.70) >consensus CACUUGGCCGUAAAUUACCAUAAAUUCCAUUCCGCAGAACAGUUUAAUAUUUAUAAUUCUAAAUCACGAGCACGACUGUCUGCAGCU______________AUAUAGUUGGCCAUAAAAA ....(((((........................(((((.((((.....(((((......)))))..((....)))))))))))((((..................)))))))))...... (-19.63 = -18.97 + -0.66)



| Location | 5,446,043 – 5,446,141 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -25.07 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5446043 98 - 27905053 ACGAGGUGUUUUAGCCAA----CUGCAGGCGCGUUCUGGCUCUAAUCUAAUC---GCUGUGGCCAUUUUGCGGUUCUGAUUUUUAUGGCCAACUAUAA--------------AGUUGCA ..((...(((..(((((.----.(((....)))...)))))..)))....))---((...((((((....((....))......))))))((((....--------------)))))). ( -22.00) >DroEre_CAF1 33194 115 - 1 ACGAGGUGUUUUAGCCAA----CUGCAGGCGCUUUCUGGCUGUAAUCUAAUCUAAGCUGUGGCUAUUUUGCGGCUCUGAUUUUUAUGGCCAACUAUGUACGAGUACAUACAAAGCUGCA ....(((......)))..----......(((((((.(((((((((...((((..(((((..(.....)..)))))..)))).)))))))))..((((((....)))))).))))).)). ( -36.50) >DroYak_CAF1 33851 105 - 1 UCGAGGUGUUUUGGCCAACUAACUGCAAGCGAGUUCUGGCUGUAAUCUAAUCUAAGCUGUGGCUAUUUUGCGGUUCUGAUUUUUAUGGCCAACCAUAU--------------AGCUGCA ....(((......)))........(((.((......(((((((((...((((..(((((..(.....)..)))))..)))).))))))))).......--------------.))))). ( -28.34) >consensus ACGAGGUGUUUUAGCCAA____CUGCAGGCGCGUUCUGGCUGUAAUCUAAUCUAAGCUGUGGCUAUUUUGCGGUUCUGAUUUUUAUGGCCAACUAUAU______________AGCUGCA ....(((......)))........(((.(((....)(((((((((...((((..(((((..(.....)..)))))..)))).)))))))))......................))))). (-25.07 = -25.30 + 0.23)

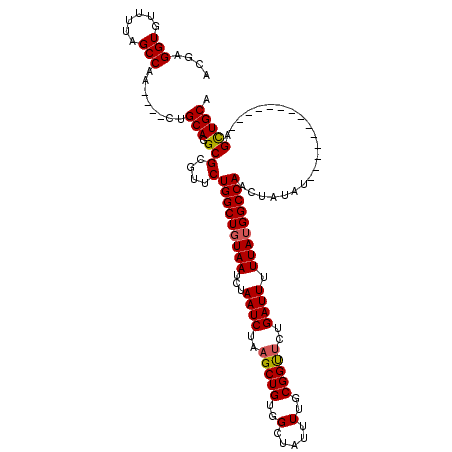

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:03 2006