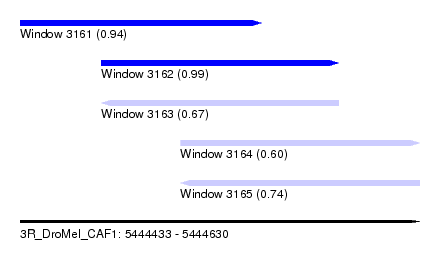
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,444,433 – 5,444,630 |
| Length | 197 |
| Max. P | 0.991388 |

| Location | 5,444,433 – 5,444,552 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.55 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -27.44 |
| Energy contribution | -28.28 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5444433 119 + 27905053 AUUUUCUGUUAAACUCUGCGGCUUGGCCUAUUGAGAACUUUCAAGUGUUUGGUCCCCA-AACAAAUAACUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACAC ......((((...(((.(((((((((((.((.(((((((.((((((((((((...)))-))).....)))).))...))))))).)).))))..........)))))))..))).)))). ( -34.10) >DroSec_CAF1 30867 119 + 1 AUUUUAUGUUCAACUCUGCGGCUUGGCCUAUUGAGAACUUUCAAGUGUUUGGUGCCCA-AACAAAUAGCUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACAC ......((((...(((.(((((((((((.((.(((((((.((((((((((((...)))-))).....)))).))...))))))).)).))))..........)))))))..))).)))). ( -34.60) >DroSim_CAF1 26084 119 + 1 AUUUUCUGUUCAACUCUGCGGCUUGGCCUAUUGAGAACUUUCAAGUGUUUGGUGCCCA-AACAAAUAGCUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACAC ......((((...(((.(((((((((((.((.(((((((.((((((((((((...)))-))).....)))).))...))))))).)).))))..........)))))))..))).)))). ( -34.40) >DroEre_CAF1 31621 119 + 1 AUUUUCUGUUCAACUCUGCGACUUGGCCUAUUGAGCACUUUCAAGUGUUUGGUCCCCAAAACAAAUAACUCUGAUGUGGUUCUCUG-CGGCUUUUGCAUUUUAAGCCGCCUGGGAAACAC ......................((((...((..((((((....))))))..))..))))................(((.(((((.(-((((((.........)))))))..))))).))) ( -30.90) >DroYak_CAF1 32237 118 + 1 AUUUUCUGCUCAACUCUGCGUCUUGGCCUAUUGAGCACUUUCAAGUGUUUGGUCGCCA-AACAAAUAACUUUGAUGUGGUUCUCUG-CGGCUUUUGCAUUUUAAGCUGCCUGGGAAACAC .......((........))((.(((((..((..((((((....))))))..)).))))-))).............(((.(((((.(-((((((.........)))))))..))))).))) ( -34.50) >consensus AUUUUCUGUUCAACUCUGCGGCUUGGCCUAUUGAGAACUUUCAAGUGUUUGGUCCCCA_AACAAAUAACUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACAC .............(((.(((((((((((....(((((((.(((((((((((..........)))))).))).))...)))))))....))))..........)))))))..)))...... (-27.44 = -28.28 + 0.84)



| Location | 5,444,473 – 5,444,590 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -30.83 |
| Energy contribution | -30.47 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5444473 117 + 27905053 UCAAGUGUUUGGUCCCCA-AACAAAUAACUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG--UUUGGGAAACAAACGCAAAUUU ....(((((((...((((-(((....((((..(((.((((..(((...(((....((.......)).)))..)))...)))).)))..))))...)--))))))...)))))))...... ( -36.20) >DroSec_CAF1 30907 117 + 1 UCAAGUGUUUGGUGCCCA-AACAAAUAGCUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGGUCUCG--UUUGGGAAACAAACGCAAAUUU ....(((((((...((((-(((....(((((.((((..(....((...))...)..))))..)))))((((((((.........))))))))...)--))))))...)))))))...... ( -43.40) >DroSim_CAF1 26124 117 + 1 UCAAGUGUUUGGUGCCCA-AACAAAUAGCUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG--UUUGGGAAACAAACGCAAAUUU ....(((((((...((((-(((...((((((.((((..(....((...))...)..))))..)))))).((((((.........)))))).....)--))))))...)))))))...... ( -39.60) >DroEre_CAF1 31661 118 + 1 UCAAGUGUUUGGUCCCCAAAACAAAUAACUCUGAUGUGGUUCUCUG-CGGCUUUUGCAUUUUAAGCCGCCUGGGAAACACCGAAUCCCAGUUCUCG-UUUUGGGAAACGAACGCAAAUUU ....(((((((...((((((((....((((..(((.((((((((.(-((((((.........)))))))..))))...)))).)))..))))...)-)))))))...)))))))...... ( -43.10) >DroYak_CAF1 32277 118 + 1 UCAAGUGUUUGGUCGCCA-AACAAAUAACUUUGAUGUGGUUCUCUG-CGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCGGCUUUGGGAAACGAACGCAAAUUU ....(((((((.((.(((-(((....((((..(((.((((((((.(-((((((.........)))))))..))))...)))).)))..))))....).)))))))..)))))))...... ( -34.70) >consensus UCAAGUGUUUGGUCCCCA_AACAAAUAACUUCGAUGUGGUUCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG__UUUGGGAAACAAACGCAAAUUU ....(((((((...((((........((((..(((.((((((((.(.((((((.........)))))).).))))...)))).)))..))))........))))...)))))))...... (-30.83 = -30.47 + -0.36)


| Location | 5,444,473 – 5,444,590 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -25.94 |
| Energy contribution | -27.74 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5444473 117 - 27905053 AAAUUUGCGUUUGUUUCCCAAA--CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGAACCACAUCGAAGUUAUUUGUU-UGGGGACCAAACACUUGA ........(((((.((((((((--((((((((..(((..(((.(((((..(((.((.......))....)))...))))))))..)))..)))).))))))-)))))).)))))...... ( -39.70) >DroSec_CAF1 30907 117 - 1 AAAUUUGCGUUUGUUUCCCAAA--CGAGACCUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGAACCACAUCGAAGCUAUUUGUU-UGGGCACCAAACACUUGA ........(((((...((((((--((((.(((((((.........))))))).(((((.....((....))....((....))......))))).))))))-))))...)))))...... ( -36.10) >DroSim_CAF1 26124 117 - 1 AAAUUUGCGUUUGUUUCCCAAA--CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGAACCACAUCGAAGCUAUUUGUU-UGGGCACCAAACACUUGA ........(((((...((((((--((((...(((..(((((((((((((.(((.((.......))....)))...)).)))..)))))))).)))))))))-))))...)))))...... ( -34.80) >DroEre_CAF1 31661 118 - 1 AAAUUUGCGUUCGUUUCCCAAAA-CGAGAACUGGGAUUCGGUGUUUCCCAGGCGGCUUAAAAUGCAAAAGCCG-CAGAGAACCACAUCAGAGUUAUUUGUUUUGGGGACCAAACACUUGA ........(((.(.(((((((((-((((((((..(((..(((.((((....(((((((.........))))))-).)))))))..)))..)))).))))))))))))).).)))...... ( -40.30) >DroYak_CAF1 32277 118 - 1 AAAUUUGCGUUCGUUUCCCAAAGCCGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCG-CAGAGAACCACAUCAAAGUUAUUUGUU-UGGCGACCAAACACUUGA ...((((((...........((((.(....((((((.........)))))).).)))).....((....))))-))))..............(((..((((-(((...)))))))..))) ( -26.50) >consensus AAAUUUGCGUUUGUUUCCCAAA__CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGAACCACAUCGAAGUUAUUUGUU_UGGGGACCAAACACUUGA ........(((((.((((((....((((((((..(((..(((.(((((..(((.((.......))....)))...))))))))..)))..)))).))))...)))))).)))))...... (-25.94 = -27.74 + 1.80)


| Location | 5,444,512 – 5,444,630 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.48 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -19.70 |
| Energy contribution | -19.66 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5444512 118 + 27905053 UCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG--UUUGGGAAACAAACGCAAAUUUAGUUGUUAACGGCCAAGAAACAAAUAAAAAAAUAGAAAAU (((((((((((.(((((............((((((.........)))))).....(--((((.....)))))))))).......))))))))...)))...................... ( -26.90) >DroSec_CAF1 30946 109 + 1 UCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGGUCUCG--UUUGGGAAACAAACGCAAAUUUAGCUGUUAACAACCAAGAAA-----AAAA----AGAAAAU (((..((((((....))......((((((((((((.........))))))))..((--((((.....)))))).......)))).....))))..)))..-----....----....... ( -29.30) >DroSim_CAF1 26163 113 + 1 UCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG--UUUGGGAAACAAACGCAAAUUUAGCUGUUAACAGCCAAGAAA-----AAAAAAGUAGAAAAU (((..((((((....))......(((((.((((((.........))))))....((--((((.....))))))......))))).....))))..)))..-----............... ( -25.40) >DroEre_CAF1 31701 111 + 1 UCUCUG-CGGCUUUUGCAUUUUAAGCCGCCUGGGAAACACCGAAUCCCAGUUCUCG-UUUUGGGAAACGAACGCAAAUUUAGCUGUUAACAGCCAAGAA-------CACAAAUACAGAAU ..((((-((((((.........))))))).(((((.........)))))(((((.(-(.((.(....).)).)).......((((....))))..))))-------)........))).. ( -30.70) >DroYak_CAF1 32316 114 + 1 UCUCUG-CGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCGGCUUUGGGAAACGAACGCAAAUUUAGCUGUUAACAGCCAAGAAA-----AAACAAAUACAAAAU .....(-((((((.........)))))))((((((.........))))))((((.(((.((.(....).)).)).......((((....))))).)))).-----............... ( -28.90) >consensus UCUCCGUUGGCUUUUGCAUUUUAAGCUGCCUGGGAAACACCGAAUCCCAGUUCUCG__UUUGGGAAACAAACGCAAAUUUAGCUGUUAACAGCCAAGAAA_____AAAAAAAUAGAAAAU ........(((((.........)))))((((((((.........))))))((((((....))))))......)).......((((....))))........................... (-19.70 = -19.66 + -0.04)

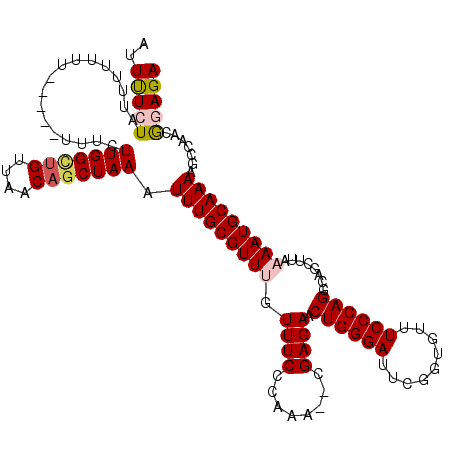
| Location | 5,444,512 – 5,444,630 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.48 |
| Mean single sequence MFE | -30.74 |
| Consensus MFE | -23.54 |
| Energy contribution | -24.22 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5444512 118 - 27905053 AUUUUCUAUUUUUUUAUUUGUUUCUUGGCCGUUAACAACUAAAUUUGCGUUUGUUUCCCAAA--CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGA ....................(((((((((.((.....))....((((((((((.....))))--)(((..((((((.........))))))....))).....))))).))))).)))). ( -26.20) >DroSec_CAF1 30946 109 - 1 AUUUUCU----UUUU-----UUUCUUGGUUGUUAACAGCUAAAUUUGCGUUUGUUUCCCAAA--CGAGACCUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGA .......----....-----..((((.((((......(((...((((((((((.....))))--)(((.(((((((.........)))))))...))).....)))))))))))).)))) ( -30.00) >DroSim_CAF1 26163 113 - 1 AUUUUCUACUUUUUU-----UUUCUUGGCUGUUAACAGCUAAAUUUGCGUUUGUUUCCCAAA--CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGA ..(((((........-----....(((((((....))))))).((((((((((.....))))--)(((..((((((.........))))))....))).....))))).......))))) ( -29.80) >DroEre_CAF1 31701 111 - 1 AUUCUGUAUUUGUG-------UUCUUGGCUGUUAACAGCUAAAUUUGCGUUCGUUUCCCAAAA-CGAGAACUGGGAUUCGGUGUUUCCCAGGCGGCUUAAAAUGCAAAAGCCG-CAGAGA .((((((.......-------...(((((((....)))))))....((.(((((((....)))-))))..((((((.........))))))))(((((.........))))))-))))). ( -36.30) >DroYak_CAF1 32316 114 - 1 AUUUUGUAUUUGUUU-----UUUCUUGGCUGUUAACAGCUAAAUUUGCGUUCGUUUCCCAAAGCCGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCG-CAGAGA .((((((((((....-----.(((((((((...(((.((.......))....)))......)))))))))((((((.........))))))........))))))))))....-...... ( -31.40) >consensus AUUUUCUAUUUUUUU_____UUUCUUGGCUGUUAACAGCUAAAUUUGCGUUUGUUUCCCAAA__CGAGAACUGGGAUUCGGUGUUUCCCAGGCAGCUUAAAAUGCAAAAGCCAACGGAGA ..(((((.................(((((((....))))))).(((((((((.((((........)))).((((((.........))))))........))))))))).......))))) (-23.54 = -24.22 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:00 2006