
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,441,960 – 5,442,114 |
| Length | 154 |
| Max. P | 0.993526 |

| Location | 5,441,960 – 5,442,074 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -31.90 |
| Consensus MFE | -24.22 |
| Energy contribution | -25.33 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5441960 114 + 27905053 GUGGCAUUGCCAUUGUUGUUGGC------GUUGCAGUUGUGCAAACGCAUACAAAACACACAUUAGGCAUCAUUUAGCAUGUUUAGCAUGUUUUGCAUUUUGCGAUUGCCCAAUUAACCC ...(((.(((((.......))))------).)))(((((.((((.((((..(((((((....((((((((........))))))))..))))))).....)))).)))).)))))..... ( -33.30) >DroEre_CAF1 29173 114 + 1 ------UUGCCAUUGUUGUUGGCGGUGCAGUUGCAGUUAUGCAAAUGCAUACAAAACACACAUUAGGCAUCAUUUUGCAUGUUUAGCAUGUUUUGCAUUUUGCUAUUGCCCAAUUAACCC ------..((((.......))))((.(((((.((((..(((((((.((((.(.((((((......)(((......))).))))).).))))))))))).)))).)))))))......... ( -33.70) >DroYak_CAF1 29714 114 + 1 ------UUGGCAUUGUUGUUGGCGUUGCAGUUGCAGUUGUGCAAACGCAUACAAAACACACAUUAGGCAUCAUUUUGCAUGUUUAGCAUGUUUUGCAUUUUGCCAUUGCCCAAUUAACCC ------........((((((((....(((((.((((..(((((((.((((.(.((((((......)(((......))).))))).).))))))))))).)))).))))))))).)))).. ( -28.70) >consensus ______UUGCCAUUGUUGUUGGCG_UGCAGUUGCAGUUGUGCAAACGCAUACAAAACACACAUUAGGCAUCAUUUUGCAUGUUUAGCAUGUUUUGCAUUUUGCCAUUGCCCAAUUAACCC ..............((((((((....(((((.((((..(((((((.((((.(.((((((......)(((......))).))))).).))))))))))).)))).))))))))).)))).. (-24.22 = -25.33 + 1.11)



| Location | 5,441,960 – 5,442,074 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.83 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -25.31 |
| Energy contribution | -26.20 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5441960 114 - 27905053 GGGUUAAUUGGGCAAUCGCAAAAUGCAAAACAUGCUAAACAUGCUAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGCACAACUGCAAC------GCCAACAACAAUGGCAAUGCCAC ((((...(((.((((.((((...((((((((((((.......((........)).....)))))))))))))))).)))).)))..))...------((((.......))))....)).. ( -33.00) >DroEre_CAF1 29173 114 - 1 GGGUUAAUUGGGCAAUAGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCAUUUGCAUAACUGCAACUGCACCGCCAACAACAAUGGCAA------ .........(((((...(((..(((((((.(((((.(((((((((.............))))))))).)))))..)))))))...)))...))).))((((.......))))..------ ( -32.82) >DroYak_CAF1 29714 114 - 1 GGGUUAAUUGGGCAAUGGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGCACAACUGCAACUGCAACGCCAACAACAAUGCCAA------ .(((..((((.....((((...(((((((((((((.......(((......))).....)))))))))))))(((.(((((....))))).)))...))))....)))))))..------ ( -34.60) >consensus GGGUUAAUUGGGCAAUAGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGCACAACUGCAACUGCA_CGCCAACAACAAUGGCAA______ ..(((.....(((.........(((((((((((((.......((........)).....)))))))))))))(((.(((((....))))).)))...)))...))).............. (-25.31 = -26.20 + 0.89)

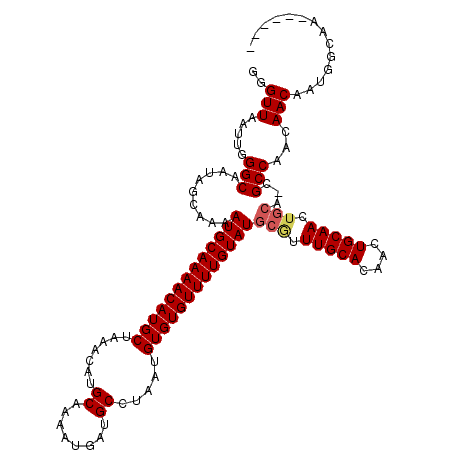
| Location | 5,441,994 – 5,442,114 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -26.86 |
| Energy contribution | -27.26 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5441994 120 - 27905053 CUUGAAAAUUUAUUAGCGCCCAGAAAAAUACCAUAUAAGCGGGUUAAUUGGGCAAUCGCAAAAUGCAAAACAUGCUAAACAUGCUAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGC ...............((((((((..((...((........)).))..))))))...((((...((((((((((((.......((........)).....))))))))))))))))...)) ( -28.90) >DroSec_CAF1 28457 120 - 1 CUUGAAAAUUUAUUAGCGCCCAGAAAAAUACCAUAUAAGCGGGUUAAUUGGGCAAUCGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGC ...............((((((((..((...((........)).))..))))))...((((...((((((((((((.......(((......))).....))))))))))))))))...)) ( -29.50) >DroSim_CAF1 23647 120 - 1 CUUGAAAAUUUAUUAGCGCCCAGAAAAAUACCAUAUAUGCGGGUUAAUUGGGCAAUCGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGC ...............((((((((..((...((........)).))..))))))...((((...((((((((((((.......(((......))).....))))))))))))))))...)) ( -29.50) >DroEre_CAF1 29207 120 - 1 CUUGCAAAUUUAUUAGCGCCAAGAAAAAUACCAUAUAAGCGGGUUAAUUGGGCAAUAGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCAUUUGC ...((((((.((((.((.((((..((....((........)).))..))))))))))(((...((((((((((((.......(((......))).....))))))))))))))))))))) ( -28.60) >DroYak_CAF1 29748 120 - 1 CUCGAAAAUUUAUUAGCGCCCAGAAAAAUACCAUAUAAGCGGGUUAAUUGGGCAAUGGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGC .................((((((..((...((........)).))..))))))....((((((((((((((((((.......(((......))).....)))))))))))))...))))) ( -28.60) >consensus CUUGAAAAUUUAUUAGCGCCCAGAAAAAUACCAUAUAAGCGGGUUAAUUGGGCAAUCGCAAAAUGCAAAACAUGCUAAACAUGCAAAAUGAUGCCUAAUGUGUGUUUUGUAUGCGUUUGC .................((((((..((...((........)).))..))))))....((((((((((((((((((.......(((......))).....)))))))))))))...))))) (-26.86 = -27.26 + 0.40)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:53 2006