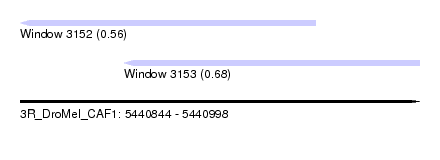
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,440,844 – 5,440,998 |
| Length | 154 |
| Max. P | 0.679140 |

| Location | 5,440,844 – 5,440,958 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.88 |
| Mean single sequence MFE | -18.22 |
| Consensus MFE | -15.44 |
| Energy contribution | -15.40 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
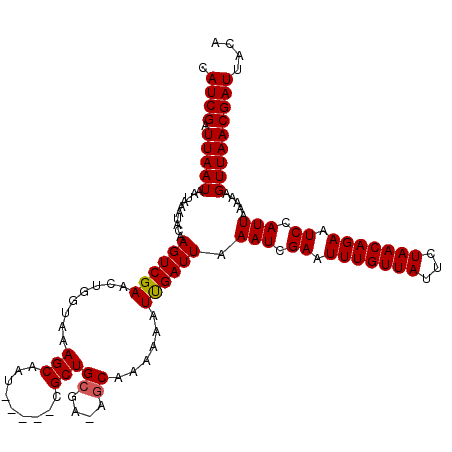
>3R_DroMel_CAF1 5440844 114 - 27905053 CAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCCAU-----CGCUGCGA-AGCAAAAAAUUGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((..(((.....))).-----.(((....-)))......)))))).(((.((.(((((((...))))))).)).))).....))))))))).... ( -18.00) >DroSec_CAF1 27326 114 - 1 CAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGA-AGCAAAAAAUCGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((...(((......))-----)(((....-)))......)))))).(((.((.(((((((...))))))).)).))).....))))))))).... ( -17.60) >DroSim_CAF1 22494 115 - 1 CAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGAAAGCAAAAAAUCGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((........(((...-----.)))((....))......)))))).(((.((.(((((((...))))))).)).))).....))))))))).... ( -21.20) >DroEre_CAF1 27981 114 - 1 CAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGA-AACAAAAAAUUGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((....((...(((..-----...)))..-.))......)))))).(((.((.(((((((...))))))).)).))).....))))))))).... ( -14.40) >DroYak_CAF1 28457 119 - 1 CAUCGAUUAAUAAUAAAUACAAGUCGAACAGGUAGAGCCAUCGCAUCGCUGCGA-AGCAAAAAAUUGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((...((.....)).(((((....)))))-.........)))))).(((.((.(((((((...))))))).)).))).....))))))))).... ( -19.90) >consensus CAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU_____CGCUGCGA_AGCAAAAAAUUGAUUAAAUCGAAUUUGUUAUUCUAACAGAAUCCAUUAAAAAGUUAACGAUUACA .((((.(((((..........((((((........(((.........)))((....))......)))))).(((.((.(((((((...))))))).)).))).....))))))))).... (-15.44 = -15.40 + -0.04)

| Location | 5,440,884 – 5,440,998 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.37 |
| Mean single sequence MFE | -23.52 |
| Consensus MFE | -18.34 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.679140 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
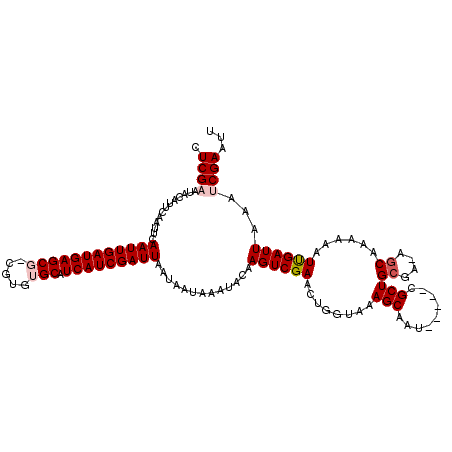
>3R_DroMel_CAF1 5440884 114 - 27905053 AUCGAAUACAUUCAAUUCAAUUGAUGAGCGUCGUGUGCAUCAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCCAU-----CGCUGCGA-AGCAAAAAAUUGAUUAAAUCGAAUU .((((.............((((((((((((.....))).))))))))).............((((((..(((.....))).-----.(((....-)))......))))))...))))... ( -24.00) >DroSec_CAF1 27366 114 - 1 CUCGAAUACAUUCAAUUCAAUUGAUGAGCGUCGUGUGCGUCAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGA-AGCAAAAAAUCGAUUAAAUCGAAUU .((((.............((((((((((((.....))).))))))))).............((((((...(((......))-----)(((....-)))......))))))...))))... ( -23.40) >DroSim_CAF1 22534 115 - 1 CUCGAAUACAUUCAAUUCAAUUGAUGAGCGGCGUGUGCAUCAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGAAAGCAAAAAAUCGAUUAAAUCGAAUU .((((.............((((((((((((.....))).))))))))).............((((((........(((...-----.)))((....))......))))))...))))... ( -26.90) >DroEre_CAF1 28021 106 - 1 CUCG-----AAUACUUCCAAUUGAUGAGC---GUGUGCAUCAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU-----CGCUGCGA-AACAAAAAAUUGAUUAAAUCGAAUU .(((-----(........(((((((((((---....)).))))))))).............((((((....((...(((..-----...)))..-.))......))))))...))))... ( -19.50) >DroYak_CAF1 28497 116 - 1 CUCGCUUCCAAUACUUCCAAUUGAUGAGC---GUGUGCAUCAUCGAUUAAUAAUAAAUACAAGUCGAACAGGUAGAGCCAUCGCAUCGCUGCGA-AGCAAAAAAUUGAUUAAAUCGAAUU ...(((((((((.......))))...(((---(.((((....((((((.............))))))...((.....))...))))))))..))-)))......(((((...)))))... ( -23.82) >consensus CUCGAAUACAUUCAAUUCAAUUGAUGAGCG_CGUGUGCAUCAUCGAUUAAUAAUAAAUACAAGUCGAACUGGUAAAGCAAU_____CGCUGCGA_AGCAAAAAAUUGAUUAAAUCGAAUU .((((.............((((((((((((.....))).))))))))).............((((((........(((.........)))((....))......))))))...))))... (-18.34 = -19.10 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:48 2006