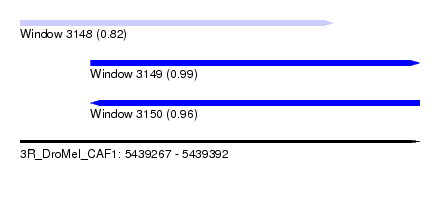
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,439,267 – 5,439,392 |
| Length | 125 |
| Max. P | 0.985472 |

| Location | 5,439,267 – 5,439,365 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -34.76 |
| Consensus MFE | -26.82 |
| Energy contribution | -27.70 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5439267 98 + 27905053 UUC------------------GCAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCC-AAAAGCCGAGCU---UUUUUAAUGGC ...------------------((.......(((.((((((..((((((((((..(....))))).(((.((....)).))).......)))))-)..)))))))))---.........)) ( -33.59) >DroSec_CAF1 25822 99 + 1 UUG------------------GCAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCGAAAAGCCGAGCU---UUUUUAAUGGC (((------------------((........((((((((.....))))((((..(....)))))..))))....((((((((.....))))))))...)))))(((---........))) ( -35.80) >DroSim_CAF1 20992 98 + 1 UUG------------------GCAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGUCUCUCUUGGCCGAAAAGCCGAGCU---UUUUUAAUGG- (((------------------((........((((((((.....))))((((..(....)))))..))))....(((((((.......)))))))...)))))...---..........- ( -35.00) >DroEre_CAF1 26411 115 + 1 UUCAGAAACUUCAGAAACUACACAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGUCGGCUCUCUUGGCCAAAAAGAA-----GUGCUUUUAAUGGC .......(((((..................(((....)))..((((((((((..(....)))))..(((((((....))).))))...))))))....)))-----))(((......))) ( -30.50) >DroYak_CAF1 26836 120 + 1 UUCAGAAACUUCACAAGCUACACAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCAAAAAGCCGAGCUGAGCUUUUAAUGGC ................((((...(((((.((((.((((((..((((((((((..(....))))).(((.((....)).))).......))))))...)))))))))))).)))...)))) ( -38.90) >consensus UUC__________________GCAAACUACAGCCUUGGCUAAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCAAAAAGCCGAGCU___UUUUUAAUGGC .................................(((((((..((((((((((..(....))))).(((.((....)).))).......))))))...)))))))................ (-26.82 = -27.70 + 0.88)

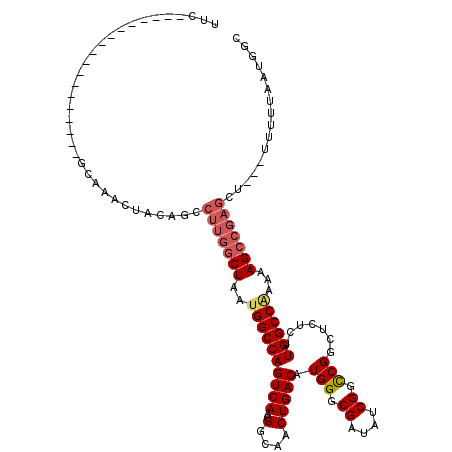
| Location | 5,439,289 – 5,439,392 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -44.28 |
| Consensus MFE | -26.70 |
| Energy contribution | -27.64 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.86 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5439289 103 + 27905053 AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCC-AAAAGCCGAGCU---UUUUUAAUGGCUCA--------GCGGCUCAGCGGCU-----GAGUGGCCUU ...((((.((((..(....)))))..((.((....)).)))))).....((((-(..(((((((((---........))))).--------((......))))))-----...))))).. ( -43.90) >DroSec_CAF1 25844 112 + 1 AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCGAAAAGCCGAGCU---UUUUUAAUGGCUCAGUGGCUCGGCGGCUCAGCGGCU-----GAGUGGCCUU ...((((.((((..(....)))))...(((....((((((((.....))))))))...)))(((((---........)))))....(((((((.((...)).)))-----)))))))).. ( -50.30) >DroSim_CAF1 21014 88 + 1 AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGUCUCUCUUGGCCGAAAAGCCGAGCU---UUUUUAAUGG------------------------CU-----GAGUGGCCUU ...(((((.((..((((..((.....))......(((((((.......)))))))...))))((((---........))------------------------))-----)).))))).. ( -35.60) >DroEre_CAF1 26451 102 + 1 AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGUCGGCUCUCUUGGCCAAAAAGAA-----GUGCUUUUAAUGGCUCAGUG--------GCUCAGUGGCU-----GAGUGGCCUU ...(((((.(((...((.(((((..(((.((....)).)))(((..((.((((((((((..-----...)))))..))))).)).)--------))))))).)))-----)).))))).. ( -37.10) >DroYak_CAF1 26876 116 + 1 AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCAAAAAGCCGAGCUGAGCUUUUAAUGGCUCAGUGGU----GAGCUCGGUGGCUGAGCUGAGUGGCCUU ...(((((.(((.((((.(((((..(((.((....)).)))((((.((((((......)))))((((((((......)))))))).).----))))))))).))))...))).))))).. ( -54.50) >consensus AAUGGCCAGUCAACGGCAACUGACAUGGGCGAUAUCGGCCGGCUCUCUUGGCCAAAAAGCCGAGCU___UUUUUAAUGGCUCAGUG________GCUCAGCGGCU_____GAGUGGCCUU ...(((((.((..((((.........((.((....)).))((((.....)))).....))))................................(((....)))......)).))))).. (-26.70 = -27.64 + 0.94)

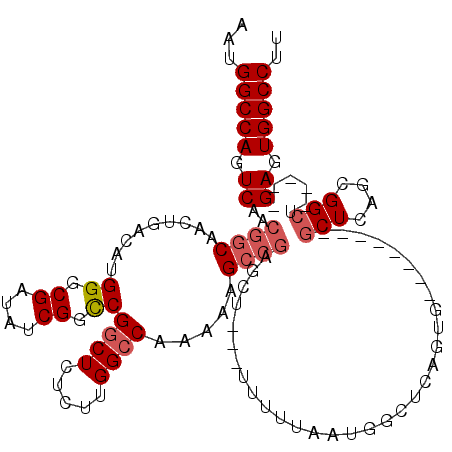
| Location | 5,439,289 – 5,439,392 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.13 |
| Mean single sequence MFE | -39.03 |
| Consensus MFE | -22.56 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5439289 103 - 27905053 AAGGCCACUC-----AGCCGCUGAGCCGC--------UGAGCCAUUAAAAA---AGCUCGGCUUUU-GGCCAAGAGAGCCGGCCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((.((-----(((.((.(.(((((--------(((((.........---.)))))))....-((((....).)))))))(((((.....)))))....)).))))).)))))... ( -41.80) >DroSec_CAF1 25844 112 - 1 AAGGCCACUC-----AGCCGCUGAGCCGCCGAGCCACUGAGCCAUUAAAAA---AGCUCGGCUUUUCGGCCAAGAGAGCCGGCCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((.((-----(((.((.(.((((((((((.................---.))))))).....((((....).)))))))(((((.....)))))....)).))))).)))))... ( -43.67) >DroSim_CAF1 21014 88 - 1 AAGGCCACUC-----AG------------------------CCAUUAAAAA---AGCUCGGCUUUUCGGCCAAGAGAGACGGCCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((.((-----((------------------------(...(((...---.((..(((...((((((.........))))))....)))...))...)))..))))).)))))... ( -30.60) >DroEre_CAF1 26451 102 - 1 AAGGCCACUC-----AGCCACUGAGC--------CACUGAGCCAUUAAAAGCAC-----UUCUUUUUGGCCAAGAGAGCCGACCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((.((-----(((......((--------(.((..((((..(((((...-----..)))))))))..)).).))((((.(((((.....))))).))))..))))).)))))... ( -31.40) >DroYak_CAF1 26876 116 - 1 AAGGCCACUCAGCUCAGCCACCGAGCUC----ACCACUGAGCCAUUAAAAGCUCAGCUCGGCUUUUUGGCCAAGAGAGCCGGCCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((.(((((.((((..(((.((((----....((((((........))))))((.((((....)))).)).)))))))..(((((.....))))).))))..))))).)))))... ( -47.70) >consensus AAGGCCACUC_____AGCCACUGAGC________CACUGAGCCAUUAAAAA___AGCUCGGCUUUUCGGCCAAGAGAGCCGGCCGAUAUCGCCCAUGUCAGUUGCCGUUGACUGGCCAUU ..(((((....................................................(((...((((((.........))))))....)))...(((((......))))))))))... (-22.56 = -23.20 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:46 2006