
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,438,590 – 5,438,722 |
| Length | 132 |
| Max. P | 0.999055 |

| Location | 5,438,590 – 5,438,709 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.74 |
| Mean single sequence MFE | -26.02 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.78 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.35 |
| SVM RNA-class probability | 0.999055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5438590 119 - 27905053 CCGGCAAAAACCGCAUGGUUAGCCCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCACAAAAAAAA-AAGGGUUCGCUUGAAUACGAAAACAAACUGAAUACAAAAGUACAUUUUUUUUG ...........(((((((.....)))))))...((((((((.......)))))))).(((((((((-..(..((((........))))..)..(((.........)))...))))))))) ( -28.90) >DroSim_CAF1 20340 111 - 1 CCGGCAAAAACCGCAUGGUGA--CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA----AAA-AAGGGAUCACUUUAAUACAAAUACAAAUUAAAUUCAAAAGUUUAUU--UUUUG ((.........(((((((...--)))))))...((((((((.......))))))))...----...-..))...................((((.(((((......)))))..--.)))) ( -22.20) >DroEre_CAF1 25784 94 - 1 CCGGCAAAAACCGCAUGGUGA--CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA----AAA-AACGGUUCACUUGAAUACAA-----------------AAUGAUCCG--UUUUU ...........(((((((...--)))))))...((((((((.......))))))))...----(((-(((((.(((.((.......)-----------------).))).)))--))))) ( -28.40) >DroYak_CAF1 26210 94 - 1 CCGGCAAAAACCGCAUGGUGA--CCAUGCAA-CGGGCUAAGAAGAAACCUUGGCUCAAA----AAAAAAGGGUUCUCUUGAAUAAAA-----------------AAUGAUUCU--UUUUU ..((......))((((((...--))))))..-.((((((((.......))))))))...----(((((((((((.(.((......))-----------------.).))))))--))))) ( -24.60) >consensus CCGGCAAAAACCGCAUGGUGA__CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA____AAA_AAGGGUUCACUUGAAUACAA_________________AAGGAUACU__UUUUG ........((((((((((.....))))))....((((((((.......))))))))..............)))).............................................. (-20.71 = -20.78 + 0.06)

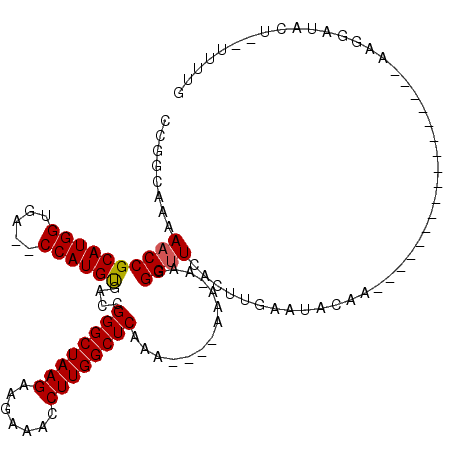
| Location | 5,438,630 – 5,438,722 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 90.13 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -21.81 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5438630 92 - 27905053 UAAAUGCAUUCAUCCGGCAAAAACCGCAUGGUUAGCCCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCACAAAAAAAA-AAGGGUUCGCUUG .....((....((((.........(((((((.....)))))))...((((((((.......))))))))..........-..))))..))... ( -24.40) >DroSim_CAF1 20378 86 - 1 UAAAUGCAUUCCUCCGGCAAAAACCGCAUGGUGA--CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA----AAA-AAGGGAUCACUUU ....((.((((((...........(((((((...--)))))))...((((((((.......))))))))...----...-.)))))))).... ( -24.50) >DroEre_CAF1 25805 86 - 1 UAAAUGCAUUCCUCCGGCAAAAACCGCAUGGUGA--CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA----AAA-AACGGUUCACUUG .............(((........(((((((...--)))))))...((((((((.......))))))))...----...-..)))........ ( -22.50) >DroYak_CAF1 26231 86 - 1 UAAAUGCAUUCCUCCGGCAAAAACCGCAUGGUGA--CCAUGCAA-CGGGCUAAGAAGAAACCUUGGCUCAAA----AAAAAAGGGUUCUCUUG ....(((.........)))..((((((((((...--))))))..-.((((((((.......))))))))...----.......))))...... ( -23.50) >consensus UAAAUGCAUUCCUCCGGCAAAAACCGCAUGGUGA__CCAUGUGACCGGGCUAAGAAGAAACCUUGGCUCAAA____AAA_AAGGGUUCACUUG ....(((.........)))..((((((((((.....))))))....((((((((.......))))))))..............))))...... (-21.81 = -21.88 + 0.06)

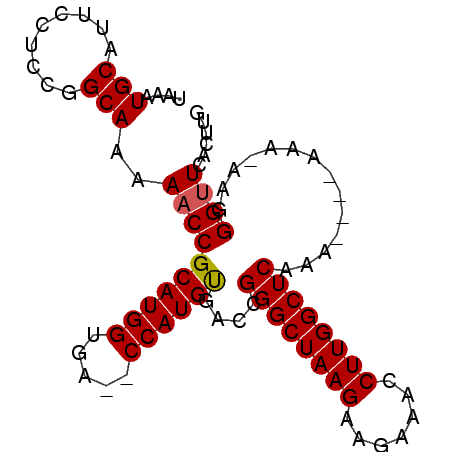
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:42 2006