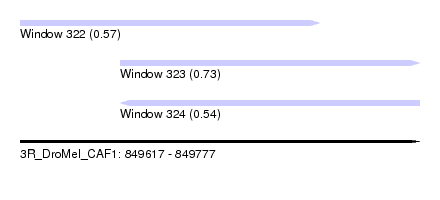
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 849,617 – 849,777 |
| Length | 160 |
| Max. P | 0.725157 |

| Location | 849,617 – 849,737 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.19 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -27.29 |
| Energy contribution | -28.35 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849617 120 + 27905053 GCUUUUUUUCAUUCUAUGUGCAACUUAUUGUUUUCCGUGUACCUUGGUGUCCGUCCGAGGAGGAACUUUCGAUUACCGAAAGGCACUUCAACCCGUUCUACGGGUCGAGGCCCCGCAGUC ((...............((((...........((((.....((((((.......)))))).))))((((((.....))))))))))(((.(((((.....))))).))).....)).... ( -35.10) >DroSec_CAF1 25308 116 + 1 GCUGUUUUUUAUUCUAUGCGAAACUGAUUGUUUUCCGUGUAGCUUGGUGU----CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGCCUCGCAGUC (((((........(((((((((((.....))))..)))))))(((((...----)))))((((..((((((.....))))))...((((.(((((.....))))).))))))))))))). ( -38.70) >DroSim_CAF1 24539 116 + 1 GCUGCUUUUUAUUCUAUGCGAAACUGAUUGUUUUCCGUGUAGCUUGGUGU----CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUAGAGGCCUCGCAGUC (((((........(((((((((((.....))))..)))))))(((((...----)))))((((..((((((.....))))))...((((.(((((.....))))).))))))))))))). ( -41.20) >DroEre_CAF1 26287 100 + 1 ----------------UGUGCCGCCGAUUGUUCUCCGUGUAGAUUGGUGU----CCGCGGAGCAAUUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGACUCGCAGUC ----------------..(((....((..((((((((((..(((....))----)))))))).)))..))((((.((....))..((((.(((((.....))))).)))))))))))... ( -38.00) >consensus GCUGUUUUUUAUUCUAUGCGAAACUGAUUGUUUUCCGUGUAGCUUGGUGU____CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGCCUCGCAGUC .........................((((((...........(((((.......)))))((((..((((((.....))))))...((((.(((((.....))))).)))))))))))))) (-27.29 = -28.35 + 1.06)

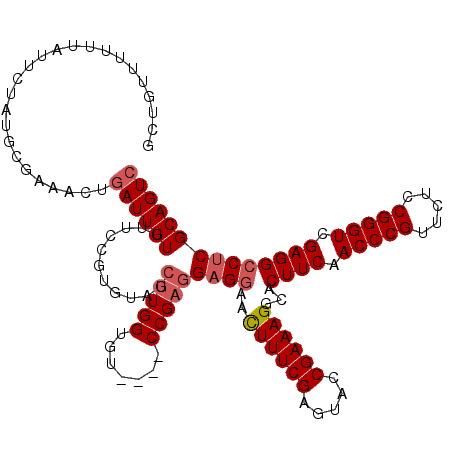
| Location | 849,657 – 849,777 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -42.92 |
| Consensus MFE | -36.59 |
| Energy contribution | -37.40 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849657 120 + 27905053 ACCUUGGUGUCCGUCCGAGGAGGAACUUUCGAUUACCGAAAGGCACUUCAACCCGUUCUACGGGUCGAGGCCCCGCAGUCCGGCUCAAACGCACACUCGGGCGUAUUCGAGUAAGACAAA ..(((((....((((((((..(((.((((((.....))))))((.((((.(((((.....))))).))))....))..))).((......))...))))))))...)))))......... ( -42.60) >DroSec_CAF1 25348 116 + 1 AGCUUGGUGU----CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGCCUCGCAGUCCGGCUCAAACGCACACUCGGGCGUAUUCGAGUAAGACAAA .(((((((((----(((((((((..((((((.....))))))...((((.(((((.....))))).))))))))...((.((.......)).)).))))))))...))))))........ ( -43.90) >DroSim_CAF1 24579 116 + 1 AGCUUGGUGU----CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUAGAGGCCUCGCAGUCCGGCUCAAACGCACACUCGGGCGUAUUCGAGUAAGACAAA .(((((((((----(((((((((..((((((.....))))))...((((.(((((.....))))).))))))))...((.((.......)).)).))))))))...))))))........ ( -43.90) >DroEre_CAF1 26311 116 + 1 AGAUUGGUGU----CCGCGGAGCAAUUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGACUCGCAGUCCGGCUCAAACGCACACUCGGGCGUAUUCGAGUAAGACAAA .......(((----(.(((((((....(((((((.((....))..((((.(((((.....))))).))))))))).))....))))...)))..(((((((....)))))))..)))).. ( -41.30) >consensus AGCUUGGUGU____CCGAGGAGGAACUUUCGAGUACCGAAAGGCACUUCAACCCGUUCUCCGGGUCGAGGCCUCGCAGUCCGGCUCAAACGCACACUCGGGCGUAUUCGAGUAAGACAAA ..(((((.......)))))((((..((((((.....))))))...((((.(((((.....))))).))))))))...(((..((((((((((........)))).)).))))..)))... (-36.59 = -37.40 + 0.81)

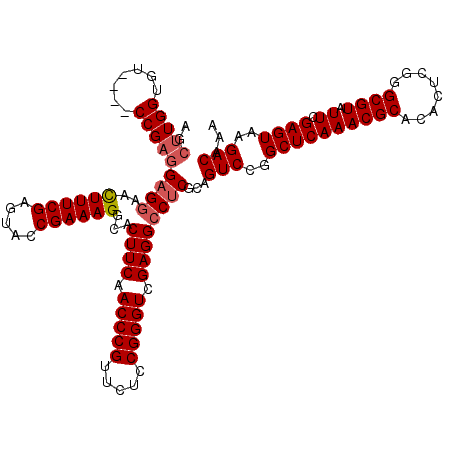
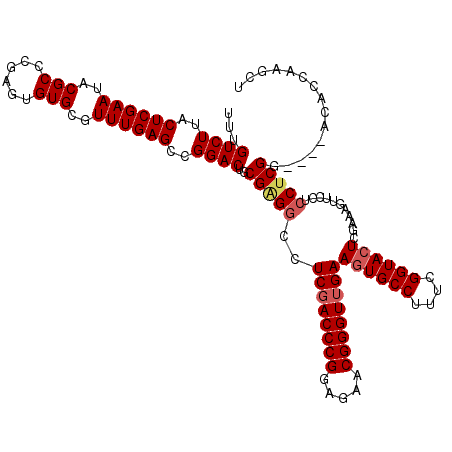
| Location | 849,657 – 849,777 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.07 |
| Mean single sequence MFE | -44.40 |
| Consensus MFE | -36.62 |
| Energy contribution | -37.50 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 849657 120 - 27905053 UUUGUCUUACUCGAAUACGCCCGAGUGUGCGUUUGAGCCGGACUGCGGGGCCUCGACCCGUAGAACGGGUUGAAGUGCCUUUCGGUAAUCGAAAGUUCCUCCUCGGACGGACACCAAGGU ..((((.((((((........))))))..((((((((..((((((..(((((((((((((.....))))))).)).))))..))))...(....)..))..))))))))))))....... ( -48.00) >DroSec_CAF1 25348 116 - 1 UUUGUCUUACUCGAAUACGCCCGAGUGUGCGUUUGAGCCGGACUGCGAGGCCUCGACCCGGAGAACGGGUUGAAGUGCCUUUCGGUACUCGAAAGUUCCUCCUCGG----ACACCAAGCU ...((..((((((........)))))).))(((((((..((((((.((((((((((((((.....))))))).)).))))).))))...(....)..))..)))))----))........ ( -44.50) >DroSim_CAF1 24579 116 - 1 UUUGUCUUACUCGAAUACGCCCGAGUGUGCGUUUGAGCCGGACUGCGAGGCCUCUACCCGGAGAACGGGUUGAAGUGCCUUUCGGUACUCGAAAGUUCCUCCUCGG----ACACCAAGCU ...((..((((((........)))))).))(((((((..((((((.((((((((.(((((.....))))).).)).))))).))))...(....)..))..)))))----))........ ( -40.70) >DroEre_CAF1 26311 116 - 1 UUUGUCUUACUCGAAUACGCCCGAGUGUGCGUUUGAGCCGGACUGCGAGUCCUCGACCCGGAGAACGGGUUGAAGUGCCUUUCGGUACUCGAAAAUUGCUCCGCGG----ACACCAAUCU ..(((((((((((........)))))).(((...((((.((((.....))))((((((((.....))))))))((((((....))))))........)))))))))----)))....... ( -44.40) >consensus UUUGUCUUACUCGAAUACGCCCGAGUGUGCGUUUGAGCCGGACUGCGAGGCCUCGACCCGGAGAACGGGUUGAAGUGCCUUUCGGUACUCGAAAGUUCCUCCUCGG____ACACCAAGCU ...((((..((((((..(((......)))..))))))..))))..(((((..((((((((.....))))))))((((((....))))))...........)))))............... (-36.62 = -37.50 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:47 2006