
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,432,968 – 5,433,123 |
| Length | 155 |
| Max. P | 0.999970 |

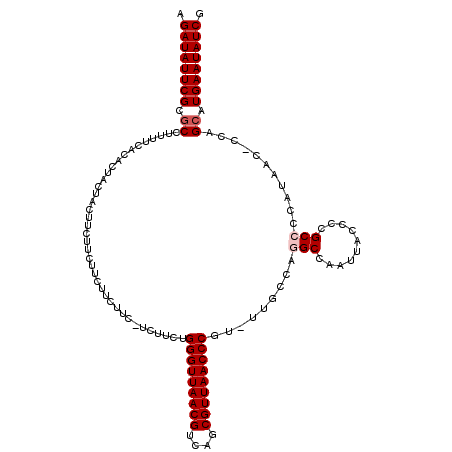
| Location | 5,432,968 – 5,433,083 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -24.78 |
| Energy contribution | -25.38 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.77 |
| SVM RNA-class probability | 0.999948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5432968 115 + 27905053 AGAUAUUCGCGCCUUUUCACACUACUACUUCUUCUUCUUGU---UCUUCUGGGUUAACGUCAGCGUUAACCCGU-UUGCCAGGCCAAUUACCCCGCCCCAUAAC-CCAGCAUGAAUAUCG .((((((((.((.............................---......(((((((((....)))))))))((-(.....(((..........)))....)))-...)).)))))))). ( -28.40) >DroSec_CAF1 19578 118 + 1 AGAUAUUCGCACCUUUUCACACUACCACUUCUUCUUCUUCUUCGUCUUCUGGGUUAACGUCAGCGUUAACCCGU-UUGCCAAGCCAAUUACCCCGCCCCAUAAC-CCAGCAUGAAUAUCG .((((((((..............................(((.((....((((((((((....)))))))))).-..)).)))...........((........-...)).)))))))). ( -23.30) >DroSim_CAF1 14609 118 + 1 AGAUAUUCGCGCCUUUUCACACUACCACUUCUUCUUCUUCUUCGUCUUCUGGGUUAACGUCAGCGUUAACCCGU-UUGCCAAGCCAAUUACCCCGCCCCAUAAC-CCAGCAUGAAUAUCG .((((((((.((......................................(((((((((....)))))))))((-(.....)))....................-...)).)))))))). ( -24.90) >DroEre_CAF1 20177 104 + 1 AGAUAUUCGCGCCUUUUCACACUACU------------UCUU---CUUCUGGGUUAACGUCAGCGUUAACCCGU-UUGCCAGGCCAAUUACCCCGCCCCAUAACCCCAGCAUGAAUAUCG .((((((((.((..............------------....---.....(((((((((....)))))))))((-(.....(((..........)))....)))....)).)))))))). ( -28.40) >DroYak_CAF1 20510 107 + 1 AGAUAUUCGCGCCUUUUCACACUACU------------UCUUCAUCUUCUGGGUUAACGUCAGCGUUAACCCGUUUUGCCAGGCCAAUUACCCCGCCCCAAAAG-CCAGCAUGAAUAUCG .((((((((.((..............------------............(((((((((....)))))))))(((((....(((..........)))...))))-)..)).)))))))). ( -30.80) >consensus AGAUAUUCGCGCCUUUUCACACUACUACUUCUUCUUCUUCUUC_UCUUCUGGGUUAACGUCAGCGUUAACCCGU_UUGCCAGGCCAAUUACCCCGCCCCAUAAC_CCAGCAUGAAUAUCG .((((((((.((......................................(((((((((....))))))))).........(((..........)))...........)).)))))))). (-24.78 = -25.38 + 0.60)


| Location | 5,432,968 – 5,433,083 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.93 |
| Mean single sequence MFE | -36.38 |
| Consensus MFE | -34.96 |
| Energy contribution | -34.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.96 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5432968 115 - 27905053 CGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCCUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGA---ACAAGAAGAAGAAGUAGUAGUGUGAAAAGGCGCGAAUAUCU .(((((((.(((((.-.((.....((.((((.....)))).))..-..(((((((((....)))))))))......---..........))..)))))((((......))))))))))). ( -37.90) >DroSec_CAF1 19578 118 - 1 CGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCUUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGACGAAGAAGAAGAAGAAGUGGUAGUGUGAAAAGGUGCGAAUAUCU .(((((((..(((..-.((((...((.((((.....)))).))..-..(((((((((....)))))))))..............................))))...)))..))))))). ( -33.00) >DroSim_CAF1 14609 118 - 1 CGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCUUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGACGAAGAAGAAGAAGAAGUGGUAGUGUGAAAAGGCGCGAAUAUCU .(((((((.(((..(-((((....((...))...)))))..))).-..(((((((((....)))))))))............................((((......))))))))))). ( -35.10) >DroEre_CAF1 20177 104 - 1 CGAUAUUCAUGCUGGGGUUAUGGGGCGGGGUAAUUGGCCUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAG---AAGA------------AGUAGUGUGAAAAGGCGCGAAUAUCU .(((((((.((((....((((...((.((((.....)))).))..-..(((((((((....))))))))).....---....------------......))))...)))).))))))). ( -37.10) >DroYak_CAF1 20510 107 - 1 CGAUAUUCAUGCUGG-CUUUUGGGGCGGGGUAAUUGGCCUGGCAAAACGGGUUAACGCUGACGUUAACCCAGAAGAUGAAGA------------AGUAGUGUGAAAAGGCGCGAAUAUCU .(((((((.((((..-((((....((.((((.....)))).)).....(((((((((....))))))))).......)))).------------))))((((......))))))))))). ( -38.80) >consensus CGAUAUUCAUGCUGG_GUUAUGGGGCGGGGUAAUUGGCCUGGCAA_ACGGGUUAACGCUGACGUUAACCCAGAAGA_GAAGAAGAAGAAGAAGUAGUAGUGUGAAAAGGCGCGAAUAUCU .(((((((.((((....((((...((.((((.....)))).)).....(((((((((....)))))))))..............................))))...)))).))))))). (-34.96 = -34.76 + -0.20)


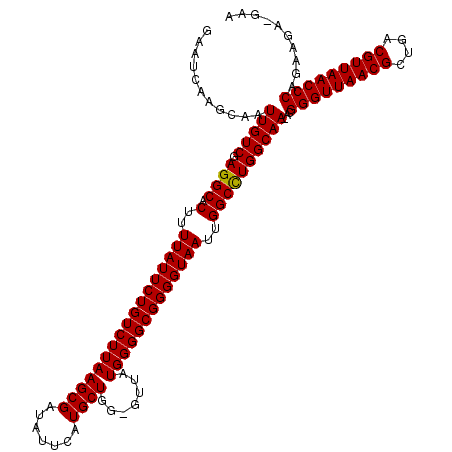
| Location | 5,433,008 – 5,433,123 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.95 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -38.53 |
| Energy contribution | -38.29 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5433008 115 - 27905053 GAAUCAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCCUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGA---A ...((......(((((.((((.(..(((((((((((((((((.......))))..-....)))))))))))))..))))))))))-..(((((((((....))))))))).))...---. ( -40.50) >DroSec_CAF1 19618 118 - 1 GAAUCAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCUUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGACGAA ...((......((((((((.((...(((((((((((((((((.......))))..-....))))))))))))).)).))))))))-..(((((((((....))))))))).......)). ( -39.70) >DroSim_CAF1 14649 118 - 1 GAAUCAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGG-GUUAUGGGGCGGGGUAAUUGGCUUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAGACGAA ...((......((((((((.((...(((((((((((((((((.......))))..-....))))))))))))).)).))))))))-..(((((((((....))))))))).......)). ( -39.70) >DroEre_CAF1 20205 116 - 1 GAAUAAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGGGGUUAUGGGGCGGGGUAAUUGGCCUGGCAA-ACGGGUUAACGCUGACGUUAACCCAGAAG---AA ...........(((((.((((.(..(((((((((((((((((.......)))).......)))))))))))))..))))))))))-..(((((((((....))))))))).....---.. ( -40.01) >DroYak_CAF1 20538 119 - 1 GAAUCAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGG-CUUUUGGGGCGGGGUAAUUGGCCUGGCAAAACGGGUUAACGCUGACGUUAACCCAGAAGAUGAA ..(((......(((((.((((.(..((((((((((((((((.(.........).)-))..)))))))))))))..))))))))))...(((((((((....)))))))))....)))... ( -42.70) >consensus GAAUCAAGCAAUUGUCGAGGCACUUUUAUUCUGUCUUAAGCGAUAUUCAUGCUGG_GUUAUGGGGCGGGGUAAUUGGCCUGGCAA_ACGGGUUAACGCUGACGUUAACCCAGAAGA_GAA ...........(((((.((((.(..(((((((((((((((((.......)))).......)))))))))))))..))))))))))...(((((((((....))))))))).......... (-38.53 = -38.29 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:34 2006