
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,428,878 – 5,428,993 |
| Length | 115 |
| Max. P | 0.777745 |

| Location | 5,428,878 – 5,428,993 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.26 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -23.22 |
| Energy contribution | -23.17 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.59 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5428878 115 + 27905053 GGUUUUUUAAUGCCCUGGAAAAAAAAGGG-GGUUGUCCAUCC----AAUAAGGUCUAUGAGAAUUUGAUUUCAUAUAAUCGAAUUCAUGGAAUUUUUUACCCCAACUUUUGGGCGCGGCC ..........(((((..(((.......((-(((..(((((((----.....)).......((((((((((......))))))))))))))).......)))))...)))..)).)))... ( -31.20) >DroSec_CAF1 15659 112 + 1 GGUUUUUUAAUGCCCUGGAAA---AGGGG-GGUUGUCCAUCC----AAUAAGGUCUAUGAGAAUUUGAUUUCAUAUAAUCGAAUUCAUGGAAUUUUUUAACCCAACUUUUGGGCGCGGCC ...........((((...(((---((..(-((((((((((((----.....)).......((((((((((......)))))))))))))))......))))))..)))))))))...... ( -33.50) >DroSim_CAF1 10641 112 + 1 GGUUUUUUAAUGCCCUGGAAA---AAGGG-GGUUGUCCAUCC----AAUAAGGUCUAUGAGAAUUUGAUUUCAUAUAAUCGAAUUCAUGGAAUUUUUUAACCCAACUUUUGGGCGCGGCC ((.....((((.((((.....---.))))-.)))).....))----.....((((.....((((((((((......))))))))))..............((((.....))))...)))) ( -30.60) >DroEre_CAF1 16078 112 + 1 GGUUUUUUAAUGCCCUGGAAAA--AAGGG-GGUUGUCCAUCC----AAUAAGGUCUAUGAGAAUUUGAUUUCUUAUAAUCGAAUUCAUGGAAUUUUU-ACCCCAACUUUGGGGCGCGGCC ...........((((..((...--...((-(((..(((((((----.....)).......((((((((((......)))))))))))))))......-)))))...))..))))...... ( -35.00) >DroYak_CAF1 16377 117 + 1 GGUUUUUUAAUGCCCUGGAAAA--AAGGGCGGUUGUCCAUCCAUUCAAUAAGGUCUAUGAGAAUUUGAUUUCUUAUAAUCGAAUUCAUGGAAUUUUU-UACCCAACUUUGGGGCGCGGCC ...........(((((......--.)))))((((((.(.((((((((..........)))((((((((((......)))))))))))))))......-..((((....))))).)))))) ( -34.30) >DroPer_CAF1 19187 96 + 1 GGUA-------GCC---GACAG-AGUGGU-AGGAGUCUGUC---------GCGUCUAUGAGAAUUUGAUUUCUUAUAAUCAGAUUCAUGGAAUUUGUGU---GUGUUUUUAGGCGCGGCU ...(-------(((---(((((-(.(...-...).))))))---------(((((((...((((((((((......))))))))))..(((((......---..)))))))))))))))) ( -34.90) >consensus GGUUUUUUAAUGCCCUGGAAAA__AAGGG_GGUUGUCCAUCC____AAUAAGGUCUAUGAGAAUUUGAUUUCAUAUAAUCGAAUUCAUGGAAUUUUUUAACCCAACUUUUGGGCGCGGCC ...........((((..((.......(((.((....)).)))...........(((((((..((((((((......)))))))))))))))...............))..))))...... (-23.22 = -23.17 + -0.05)

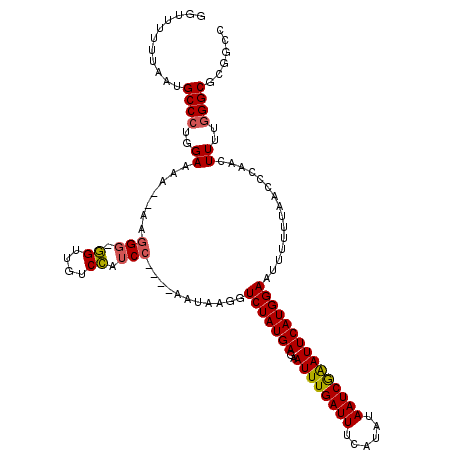
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:26 2006