
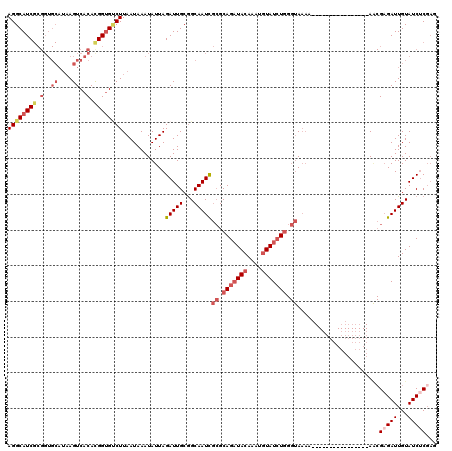
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,421,901 – 5,422,302 |
| Length | 401 |
| Max. P | 0.975667 |

| Location | 5,421,901 – 5,422,005 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.89 |
| Mean single sequence MFE | -27.54 |
| Consensus MFE | -16.68 |
| Energy contribution | -19.08 |
| Covariance contribution | 2.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421901 104 + 27905053 AGGCAUCGCGGUGCAUAAGUCACACGGUGUCUUAAUAAAUAUUAGAUUGCGGCAAUUGCGCAGAUACAAAUGUAUCUGGGUAAAA----------------AGCGAGAUUGUAUCUCCAG ((((((((..((((....).))).))))))))................((.....((((.(((((((....))))))).))))..----------------.))(((((...)))))... ( -29.90) >DroPse_CAF1 8770 85 + 1 AGGCAUC----------AGUCACACGGAGACUUAAUAAAUAUUAGAUUGCGGCAAUCCAGCAGAUACAUACAUAUGU----A-CA----------------A--GCGAUUGUAUCUCG-- .(((...----------.))).....((((.(((((....)))))......((((((..((...(((((....))))----)-..----------------.--)))))))).)))).-- ( -14.40) >DroSec_CAF1 8577 104 + 1 AGGCAUCGCGGUGCAUAAGUCACACGGUGUCUUAAUAAAUAUUAGAUUGCGGCAAUCGCGCAGCUACAAAUGUAUCUGGGUAAAA----------------AGCGAGAUUGUAUCUCGAG ((((((((..((((....).))).))))))))............(((((...)))))((.(((.(((....))).))).))....----------------..((((((...)))))).. ( -27.00) >DroEre_CAF1 9090 104 + 1 AGGCAUCGCGGUGCAUAAGUCACAUGGUGUCUUAAUAAAUAUUAGAUUGCAGCAAUCGCGCAGAUACAAAUGUAUCUGGGUAAAA----------------AACGAGAUUGUAUCUCGAG ((((((((..((((....).))).))))))))............(((((...)))))((.(((((((....))))))).))....----------------..((((((...)))))).. ( -30.10) >DroYak_CAF1 9307 120 + 1 AGGCAUCGCGGUGCAUAAGUCACACGGUGCCUUAAUAAAUAUUAGAUUGCGGCAAUCGCGCAGAUACAAAUGUAUCUGGGUAAAAACUAAAAAAAAAAAAGAACGAGAUUGUAUCGCGAG ((((((((..((((....).))).))))))))..............(((((((((((.(((((((((....)))))))(((....)))...............)).))))))..))))). ( -36.30) >consensus AGGCAUCGCGGUGCAUAAGUCACACGGUGUCUUAAUAAAUAUUAGAUUGCGGCAAUCGCGCAGAUACAAAUGUAUCUGGGUAAAA________________AACGAGAUUGUAUCUCGAG ((((((((.(..((....))..).))))))))............(((((...)))))((.(((((((....))))))).)).......................(((((...)))))... (-16.68 = -19.08 + 2.40)


| Location | 5,421,981 – 5,422,078 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -23.30 |
| Energy contribution | -23.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421981 97 + 27905053 UAAAA----------------AGCGAGAUUGUAUCUCCAGUCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUC .....----------------.((.(((((((((...(((((((((.(((((.(((.(((((.....)))))...))--))))))....)))))))))...)))))))))))... ( -28.00) >DroSec_CAF1 8657 99 + 1 UAAAA----------------AGCGAGAUUGUAUCUCGAGUCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUGUGUUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUC .....----------------.((.((((((((.((.(((((((((.(((((((((.(((((.....)))))...)))).)))))....)))))))).).))))))))))))... ( -28.60) >DroEre_CAF1 9170 97 + 1 UAAAA----------------AACGAGAUUGUAUCUCGAGUCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUC .....----------------.((.((((((((.((.(((((((((.(((((.(((.(((((.....)))))...))--))))))....)))))))).).))))))))))))... ( -24.90) >DroYak_CAF1 9387 113 + 1 UAAAAACUAAAAAAAAAAAAGAACGAGAUUGUAUCGCGAGUCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUC ......................((.(((((((((.((.((((((((.(((((.(((.(((((.....)))))...))--))))))....))))))))))..)))))))))))... ( -26.90) >consensus UAAAA________________AACGAGAUUGUAUCUCGAGUCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG__UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUC ......................((.(((((((((....((((((((.((((((....))((((.....)))).........))))....))))))))....)))))))))))... (-23.30 = -23.05 + -0.25)



| Location | 5,421,981 – 5,422,078 |
|---|---|
| Length | 97 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 89.94 |
| Mean single sequence MFE | -23.15 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.87 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.719144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421981 97 - 27905053 GAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGACUGGAGAUACAAUCUCGCU----------------UUUUA ........(((((((((....)))))))))......--.(((((((..(((.....((....))..(((.(((.....))).)))..)))))))----------------))).. ( -22.60) >DroSec_CAF1 8657 99 - 1 GAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAACACAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGACUCGAGAUACAAUCUCGCU----------------UUUUA ........(((((((((....))))))))).........(((((((..(((.....((....))..(((.((((...)))).)))..)))))))----------------))).. ( -23.30) >DroEre_CAF1 9170 97 - 1 GAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGACUCGAGAUACAAUCUCGUU----------------UUUUA ........(((((((((....)))))))))..((((--(...((((((.(.......).)))))).(((.((((...)))).)))......)))----------------))... ( -24.40) >DroYak_CAF1 9387 113 - 1 GAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGACUCGCGAUACAAUCUCGUUCUUUUUUUUUUUUAGUUUUUA ........(((((((((....)))))))))..((((--.(((((.((.(((......((((.(.(((.....)))).))))......))).)).))))).))))........... ( -22.30) >consensus GAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA__CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGACUCGAGAUACAAUCUCGCU________________UUUUA (((.....(((((((((....)))))))))............((((((.(.......).)))))).(((.(((.....))).))).))).......................... (-20.62 = -20.87 + 0.25)


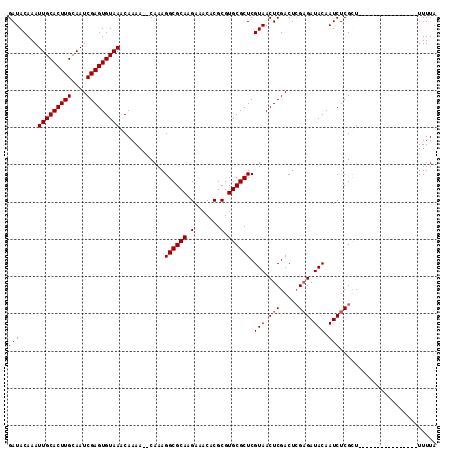
| Location | 5,422,005 – 5,422,116 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -32.60 |
| Consensus MFE | -30.39 |
| Energy contribution | -30.20 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.896538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422005 111 + 27905053 UCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUCUGGACGGAUACAUUCCAUGCCAC--ACAUGUGCUUUGCAU ((((((.(((((.(((.(((((.....)))))...))--))))))....)))))).((((((.(((...(((((((....)))))))...((((....--.))))))))))))). ( -32.40) >DroSec_CAF1 8681 114 + 1 UCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUGUGUUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUCUGG-CGGAUACAUUCCAUGCCACACACAUGUGCUUUGCAU ((((((.(((((((((.(((((.....)))))...)))).)))))....)))))).((((((.(((...(((.(.(((-((((.....)))..)))).).)))..))))))))). ( -37.30) >DroEre_CAF1 9194 108 + 1 UCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUCUGG-CGGAUACAUUCCAUGCCA----CAUGUGCUUUGCAU ((((((.(((((.(((.(((((.....)))))...))--))))))....)))))).((((((.(((...(((....((-((((.....)))..))))----))..))))))))). ( -31.10) >DroYak_CAF1 9427 108 + 1 UCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG--UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUCUGG-CAGAUACAUUCCAUGCCA----CAUGUGCUUUGCAU ((((((.(((((.(((.(((((.....)))))...))--))))))....)))))).((((((.(((...((((((...-..))))))...((((...----))))))))))))). ( -29.60) >consensus UCGAGUUACGAGCGCACGCGUGUUUCUUGCGCCUUUG__UUUUGUUUACACUCGAUUGCAAGUGCAAUUUGUAUCUGG_CGGAUACAUUCCAUGCCA____CAUGUGCUUUGCAU ((((((.((((((....))((((.....)))).........))))....)))))).((((((.(((...(((((((....)))))))...((((.......))))))))))))). (-30.39 = -30.20 + -0.19)



| Location | 5,422,005 – 5,422,116 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.14 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -27.80 |
| Energy contribution | -28.30 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422005 111 - 27905053 AUGCAAAGCACAUGU--GUGGCAUGGAAUGUAUCCGUCCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGA .(((..(((.(((((--(((...((...((((((......))))))..(((((((((....))))))))).))...--......(....)...)))))))).))).)))...... ( -32.30) >DroSec_CAF1 8681 114 - 1 AUGCAAAGCACAUGUGUGUGGCAUGGAAUGUAUCCG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAACACAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGA .(((..(((.(((((((((.((..(((.....)))(-((........(((((....)))))...((((.......))))...))))).....))))))))).))).)))...... ( -35.20) >DroEre_CAF1 9194 108 - 1 AUGCAAAGCACAUG----UGGCAUGGAAUGUAUCCG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGA .(((..(((.((((----((...((...((((((..-...))))))..(((((((((....))))))))).))...--....(.(....)...).)))))).))).)))...... ( -27.90) >DroYak_CAF1 9427 108 - 1 AUGCAAAGCACAUG----UGGCAUGGAAUGUAUCUG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA--CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGA .(((..(((.((((----((...((...((((((..-...))))))..(((((((((....))))))))).))...--....(.(....)...).)))))).))).)))...... ( -28.60) >consensus AUGCAAAGCACAUG____UGGCAUGGAAUGUAUCCG_CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA__CAAAGGCGCAAGAAACACGCGUGCGCUCGUAACUCGA .(((((.((((((((.....)))))...((((((......))))))...)))..))))).(((((((....)..........((((((.(.......).))))))....)))))) (-27.80 = -28.30 + 0.50)

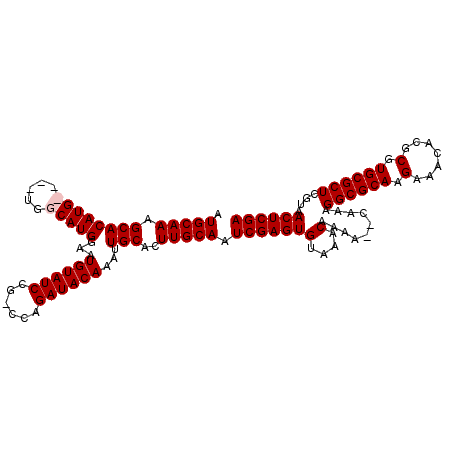
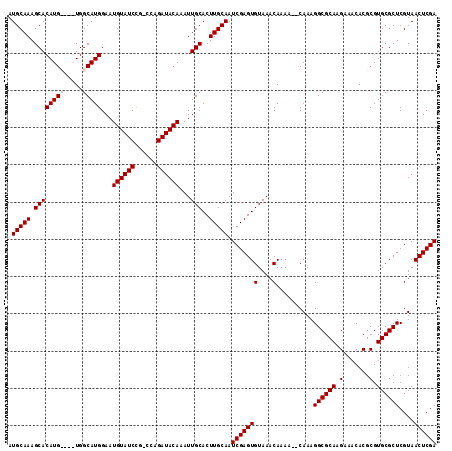
| Location | 5,422,042 – 5,422,153 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 95.10 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -27.20 |
| Energy contribution | -26.95 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422042 111 - 27905053 CAGUUAAGCCAAGUUGGCCAUUUGAGCAGGCCUUUU-UAUGCAAAGCACAUGU--GUGGCAUGGAAUGUAUCCGUCCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA- ..(((..(((.....)))((((((((((((...(((-(((((...(((....)--)).))))))))((((((......)))))).......)))))..)))))))..)))....- ( -31.30) >DroSec_CAF1 8719 114 - 1 CAGUUAAGCCAAGUUGGCCAUUUGAGCGGGCCUUUUAUAUGCAAAGCACAUGUGUGUGGCAUGGAAUGUAUCCG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAAC .......(((.....))).(((((.(((((.......(((((...((((....)))).))))).......))))-)))))).....(((((((((....)))))))))....... ( -34.84) >DroEre_CAF1 9231 108 - 1 CAGUUAAGCCGAGUUGGCCAUUUGAGCGGGCCUUUU-UAUGCAAAGCACAUG----UGGCAUGGAAUGUAUCCG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA- .......(((.....))).(((((.(((((...(((-(((((...((....)----).))))))))....))))-)))))).....(((((((((....)))))))))......- ( -32.30) >DroYak_CAF1 9464 108 - 1 CAGUUAAGCCGAGUUGGCCAUUUGAGCGGGCCUUUU-UAUGCAAAGCACAUG----UGGCAUGGAAUGUAUCUG-CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA- .......(((.....))).(((((.(((((...(((-(((((...((....)----).))))))))....))))-)))))).....(((((((((....)))))))))......- ( -30.10) >consensus CAGUUAAGCCAAGUUGGCCAUUUGAGCGGGCCUUUU_UAUGCAAAGCACAUG____UGGCAUGGAAUGUAUCCG_CCAGAUACAAAUUGCACUUGCAAUCGAGUGUAAACAAAA_ .......((((....((((.........)))).......(((...)))........))))......((((((......))))))..(((((((((....)))))))))....... (-27.20 = -26.95 + -0.25)



| Location | 5,422,078 – 5,422,193 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -34.22 |
| Consensus MFE | -26.96 |
| Energy contribution | -27.52 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422078 115 + 27905053 UGGACGGAUACAUUCCAUGCCAC--ACAUGUGCUUUGCAUA-AAAAGGCCUGCUCAAAUGGCCAACUUGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCA .....(((.....)))..((((.--...(((((...)))))-....((((.........))))....))))......(((.(((((((..((((....))))..))))))).)))... ( -33.90) >DroSec_CAF1 8756 117 + 1 UGG-CGGAUACAUUCCAUGCCACACACAUGUGCUUUGCAUAUAAAAGGCCCGCUCAAAUGGCCAACUUGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCA ..(-((((.....)))..((((.....((((((...))))))....((((.........))))....))))......(((.(((((((..((((....))))..))))))).))))). ( -35.80) >DroEre_CAF1 9267 105 + 1 UGG-CGGAUACAUUCCAUGCCA----CAUGUGCUUUGCAUA-AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACA-------UCUGCUAGAUACGCA ..(-((((.....)))..(((.----..(((((...)))))-....((((.........)))).....)))......(((.(((((((((....))-------.))))))).))))). ( -32.60) >DroYak_CAF1 9500 112 + 1 UGG-CAGAUACAUUCCAUGCCA----CAUGUGCUUUGCAUA-AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACAUUUUGUAUCUGCUAGAUACGCA (((-((((((((......(((.----..(((((...)))))-....((((.........)))).....)))...(((((.....)))))..........)))))))))))........ ( -34.60) >consensus UGG_CGGAUACAUUCCAUGCCA____CAUGUGCUUUGCAUA_AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCA (((.(((((((.......(((.......(((((...))))).....((((.........)))).....)))...(((((.....)))))...........))))))))))........ (-26.96 = -27.52 + 0.56)


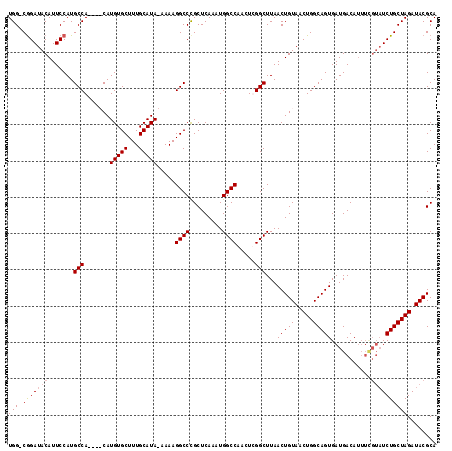
| Location | 5,422,116 – 5,422,229 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -35.15 |
| Consensus MFE | -22.42 |
| Energy contribution | -23.68 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422116 113 + 27905053 A-AAAAGGCCUGCUCAAAUGGCCAACUUGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCAGAUACACAGCU----AUCUGGCGGAUAGCUGAAGCGUAAA .-....((((.........))))......((((.(((((.....)))))...........((((((((.......)))))))).(((((----(((.....))))))))))))..... ( -40.00) >DroSec_CAF1 8795 114 + 1 AUAAAAGGCCCGCUCAAAUGGCCAACUUGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCAGCUACACAGCU----AUCUGGCGGAUAGCUGAAGCGUAAA ......((((.........))))......((((....(((.(((((((..((((....))))..))))))).))).........(((((----(((.....))))))))))))..... ( -37.50) >DroEre_CAF1 9302 110 + 1 A-AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACA-------UCUGCUAGAUACGCAGAUACACAGCUAAAUAUCUGGCGGAUGGCUCAAGCGUAAA .-....((((.........))))......((((.(((((.....)))))((...((-------(((((((((((.((.........))....)))))))))))))..))))))..... ( -33.90) >DroYak_CAF1 9535 103 + 1 A-AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACAUUUUGUAUCUGCUAGAUACGCAGAUACACAAACACA--------------GCCAUGCGUAAA .-....((((.........)))).....((((..(((((.....)))))..........(((((((((.......))))))))).......)--------------)))......... ( -29.20) >consensus A_AAAAGGCCCGCUCAAAUGGCCAACUCGGCUUAACUGUAACUGGCAGUGAUGACAUUUCGUAUCUGCUAGAUACGCAGAUACACAGCU____AUCUGGCGGAUAGCUCAAGCGUAAA ......((((.........))))......((((.(((((.....)))))...........((((((((.......))))))))..........................))))..... (-22.42 = -23.68 + 1.25)


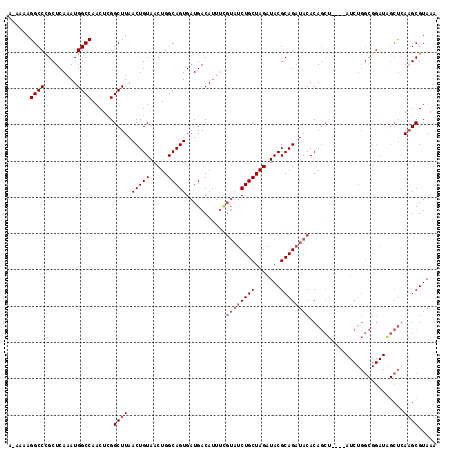
| Location | 5,422,116 – 5,422,229 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 83.55 |
| Mean single sequence MFE | -32.91 |
| Consensus MFE | -20.46 |
| Energy contribution | -22.15 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422116 113 - 27905053 UUUACGCUUCAGCUAUCCGCCAGAU----AGCUGUGUAUCUGCGUAUCUAGCAGAUACGAAAUGUCAUCACUGCCAGUUACAGUUAAGCCAAGUUGGCCAUUUGAGCAGGCCUUUU-U .........((((((((.....)))----)))))(((((((((.......))))))))).............(((.(((.(((....(((.....)))...)))))).))).....-. ( -34.40) >DroSec_CAF1 8795 114 - 1 UUUACGCUUCAGCUAUCCGCCAGAU----AGCUGUGUAGCUGCGUAUCUAGCAGAUACGAAAUGUCAUCACUGCCAGUUACAGUUAAGCCAAGUUGGCCAUUUGAGCGGGCCUUUUAU ....(((((((((((((.....)))----)))))((((((((((((((.....))))))...((....))....)))))))).....(((.....))).....))))).......... ( -34.90) >DroEre_CAF1 9302 110 - 1 UUUACGCUUGAGCCAUCCGCCAGAUAUUUAGCUGUGUAUCUGCGUAUCUAGCAGA-------UGUCAUCACUGCCAGUUACAGUUAAGCCGAGUUGGCCAUUUGAGCGGGCCUUUU-U ....((((..(((((((.(((((((((........))))))).(((.((.((((.-------((....)))))).)).)))......)).))..))))...)..))))........-. ( -31.50) >DroYak_CAF1 9535 103 - 1 UUUACGCAUGGC--------------UGUGUUUGUGUAUCUGCGUAUCUAGCAGAUACAAAAUGUCAUCACUGCCAGUUACAGUUAAGCCGAGUUGGCCAUUUGAGCGGGCCUUUU-U ....(((.((((--------------.(((....(((((((((.......))))))))).........))).))))....(((....(((.....)))...))).)))........-. ( -30.82) >consensus UUUACGCUUCAGCUAUCCGCCAGAU____AGCUGUGUAUCUGCGUAUCUAGCAGAUACGAAAUGUCAUCACUGCCAGUUACAGUUAAGCCAAGUUGGCCAUUUGAGCGGGCCUUUU_U ....(((((((..................((((((((((((((.......))))))))....((.((....)).))...))))))..(((.....)))...))))))).......... (-20.46 = -22.15 + 1.69)



| Location | 5,422,193 – 5,422,302 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -28.82 |
| Consensus MFE | -6.26 |
| Energy contribution | -5.46 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5422193 109 - 27905053 AAUUGAUAAAUAUUAAUAAGGCCAGAGGGUUAUUUGU---CGUAUUUGUUUUUGCU-GCU--AG-CGUGCGUGCAUUGCAUUUACGCUUCAGCUAUCCGCCAGAU---AGCUGUGUAUC ....((((((((........(((....))))))))))---)(((..(((...((((-((.--..-...))).)))..)))..)))((..((((((((.....)))---))))).))... ( -30.70) >DroPse_CAF1 8996 107 - 1 AAUUGAUAAAUAUUAAUAAGGCCAGAAGAGUGUGUGUAUCGGCAUCUGUUUGUGAUUGUGCGAUUGCUGCGUGCAUUGCAUUUACG-------UAUCUGGCAGAU---ACUCGUACA-- .((((((....))))))...((((((...(((.(((((...((((..((..(..(((....)))..).))))))..))))).))).-------..))))))....---.........-- ( -27.50) >DroSec_CAF1 8873 108 - 1 AAUUGAUAAAUAUUAAUAAGG-CAGAGAGUUAUUUGU---CGUAUUUGUUUUUGCU-GCU--AG-CGUGCGUGCAUUGCAUUUACGCUUCAGCUAUCCGCCAGAU---AGCUGUGUAGC ....(((((((((......((-((((......)))))---))))))))))...(((-((.--((-((((.((((...)))).)))))).((((((((.....)))---))))).))))) ( -34.50) >DroYak_CAF1 9612 99 - 1 AAUUGAUAAAUAUUAAUAAGGCCAGAGGGUUAUUUGU---CGCAUUUGUUUUUGCU-GUG--AG-CGUGCGUGCAUUGCAUUUACGCAUGGC-------------UGUGUUUGUGUAUC .........((((.((((.(((((............(---((((...((....)))-)))--)(-((((.((((...)))).))))).))))-------------).)))).))))... ( -24.40) >DroPer_CAF1 9511 107 - 1 AAUUGAUAAAUAUUAAUAAGGCCAGAAGAGUGUGUGUAUCGGCAUCUGUUUGUGAUUGUGCGAUUGGUGCGUGCAUUGCAUUUACG-------UAUCUGGCAGAU---ACUCGUACA-- .((((((....))))))...((((((...(((.(((((.(((((((...((((......))))..))))).))...))))).))).-------..))))))....---.........-- ( -27.00) >consensus AAUUGAUAAAUAUUAAUAAGGCCAGAGGGUUAUUUGU___CGCAUUUGUUUUUGCU_GUG__AG_CGUGCGUGCAUUGCAUUUACGC_U____UAUCCGCCAGAU___ACUUGUGUA_C .((((((....))))))..........((((..........(((........)))...........(((.((((...)))).))).......................))))....... ( -6.26 = -5.46 + -0.80)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:19 2006