
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,421,776 – 5,421,886 |
| Length | 110 |
| Max. P | 0.999225 |

| Location | 5,421,776 – 5,421,866 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -20.19 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421776 90 + 27905053 CCUGAAAGGUAAACAAUGUCGGGAGACACAUGUAUCUGAG-CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAUACAUACAUGUAGGUACG ((((............((((....))))(((((((.....-...(((((((((((.(.....).))))))))))).))))))))))).... ( -26.70) >DroSec_CAF1 8465 77 + 1 CCUGAAAGGUAAACAAUGUCGGGAGACACAUGUAUC--------UAUCUGGUGACCCAAUUUGAGUCAUCAGAUACAU------GGGCACG ((((..(((((.....((((....))))....))))--------)((((((((((.(.....).)))))))))).)).------))..... ( -24.00) >DroEre_CAF1 8979 76 + 1 CCUGAAAGGUAAACAAUGUCGGGAGACAUAU----CUGAG-CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU----------GGGUACG ..((..(((((.....((((....)))))))----))...-))((((((((((((.(.....).))))))))))----------))..... ( -25.60) >DroYak_CAF1 9195 77 + 1 CCUGAAAGGUAAACAAUGUCGGGAGACAUAU----CUGAGCCACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU----------GGGUACG .......(((.....(((((....)))))..----....))).((((((((((((.(.....).))))))))))----------))..... ( -27.20) >consensus CCUGAAAGGUAAACAAUGUCGGGAGACACAU____CUGAG_CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU__________GGGUACG (((.............((((....)))).................((((((((((.(.....).))))))))))..........))).... (-20.19 = -20.00 + -0.19)



| Location | 5,421,776 – 5,421,866 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 83.75 |
| Mean single sequence MFE | -22.55 |
| Consensus MFE | -17.51 |
| Energy contribution | -17.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975489 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421776 90 - 27905053 CGUACCUACAUGUAUGUAUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG-CUCAGAUACAUGUGUCUCCCGACAUUGUUUACCUUUCAGG ....(((((((((((.((((((.((((((.....)))))).))))))(..-..)..))))))))(((....)))..............))) ( -25.10) >DroSec_CAF1 8465 77 - 1 CGUGCCC------AUGUAUCUGAUGACUCAAAUUGGGUCACCAGAUA--------GAUACAUGUGUCUCCCGACAUUGUUUACCUUUCAGG .....((------((.((((((.((((((.....)))))).))))))--------.))(((.(((((....))))))))..........)) ( -22.10) >DroEre_CAF1 8979 76 - 1 CGUACCC----------AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG-CUCAG----AUAUGUCUCCCGACAUUGUUUACCUUUCAGG .((((..----------(((((.((((((.....)))))).))))).)))-)..((----(((((((....)))).))))).......... ( -22.40) >DroYak_CAF1 9195 77 - 1 CGUACCC----------AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUGGCUCAG----AUAUGUCUCCCGACAUUGUUUACCUUUCAGG .(..(((----------(((((.((((((.....)))))).))))).).))..)((----(((((((....)))).))))).......... ( -20.60) >consensus CGUACCC__________AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG_CUCAG____AUAUGUCUCCCGACAUUGUUUACCUUUCAGG .(((.............(((((.((((((.....)))))).)))))................(((((....)))))....)))........ (-17.51 = -17.07 + -0.44)



| Location | 5,421,793 – 5,421,886 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -26.75 |
| Energy contribution | -26.38 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -4.45 |
| Structure conservation index | 0.80 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421793 93 + 27905053 GUCGGGAGACACAUGUAUCUGAG-CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAUACAUACAUGUAGGUACGACAAUUGCAUCCCGGCUAAU (((((((..((..((((((((..-...(((((((((((.(.....).)))))))))))........)))))))).....))..))))))).... ( -33.82) >DroSec_CAF1 8482 80 + 1 GUCGGGAGACACAUGUAUC--------UAUCUGGUGACCCAAUUUGAGUCAUCAGAUACAU------GGGCACGACAAUUGCAUCCCGGCGAAU (((((((....((((((..--------..(((((((((.(.....).))))))))))))))------).(((.......))).))))))).... ( -32.70) >DroEre_CAF1 8996 79 + 1 GUCGGGAGACAUAU----CUGAG-CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU----------GGGUACGACAAUUGCAUCCCGACGAAA (((((((..((...----.((.(-(.((((((((((((.(.....).))))))))))----------)))).)).....))..))))))).... ( -34.20) >DroYak_CAF1 9212 80 + 1 GUCGGGAGACAUAU----CUGAGCCACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU----------GGGUACGACAAUUGCAUCCCGGCGAAA (((((((..((...----........((((((((((((.(.....).))))))))))----------))..........))..))))))).... ( -32.20) >consensus GUCGGGAGACACAU____CUGAG_CACUAUCUGGUGACCCAAUUUGAGUCAUCAGAU__________GGGUACGACAAUUGCAUCCCGGCGAAA (((((((.....................((((((((((.(.....).))))))))))............(((.......))).))))))).... (-26.75 = -26.38 + -0.38)

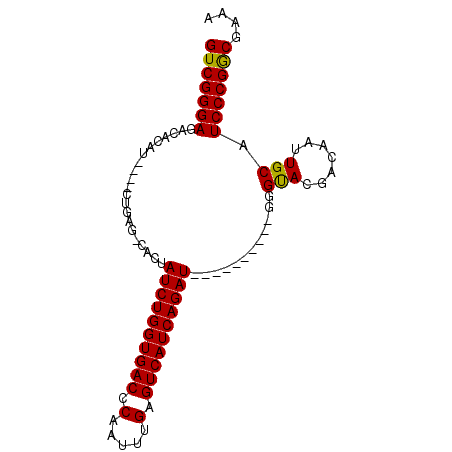
| Location | 5,421,793 – 5,421,886 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -22.17 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.78 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421793 93 - 27905053 AUUAGCCGGGAUGCAAUUGUCGUACCUACAUGUAUGUAUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG-CUCAGAUACAUGUGUCUCCCGAC ......(((((.(((..((((((....))..((((.((((((.((((((.....)))))).)))))))))-)...))))....))).))))).. ( -30.90) >DroSec_CAF1 8482 80 - 1 AUUCGCCGGGAUGCAAUUGUCGUGCCC------AUGUAUCUGAUGACUCAAAUUGGGUCACCAGAUA--------GAUACAUGUGUCUCCCGAC ......(((((.(((.....((((...------((.((((((.((((((.....)))))).))))))--------.)).))))))).))))).. ( -28.60) >DroEre_CAF1 8996 79 - 1 UUUCGUCGGGAUGCAAUUGUCGUACCC----------AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG-CUCAG----AUAUGUCUCCCGAC ....(((((((.((((((...((((..----------(((((.((((((.....)))))).))))).)))-)...)----)).))).))))))) ( -33.40) >DroYak_CAF1 9212 80 - 1 UUUCGCCGGGAUGCAAUUGUCGUACCC----------AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUGGCUCAG----AUAUGUCUCCCGAC ......(((((.(((..(((((..(((----------(((((.((((((.....)))))).))))).).))..).)----)))))).))))).. ( -29.30) >consensus AUUCGCCGGGAUGCAAUUGUCGUACCC__________AUCUGAUGACUCAAAUUGGGUCACCAGAUAGUG_CUCAG____AUAUGUCUCCCGAC ......(((((.(((...(.....)............(((((.((((((.....)))))).))))).................))).))))).. (-22.17 = -22.18 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:10 2006