
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 5,421,155 – 5,421,293 |
| Length | 138 |
| Max. P | 0.908515 |

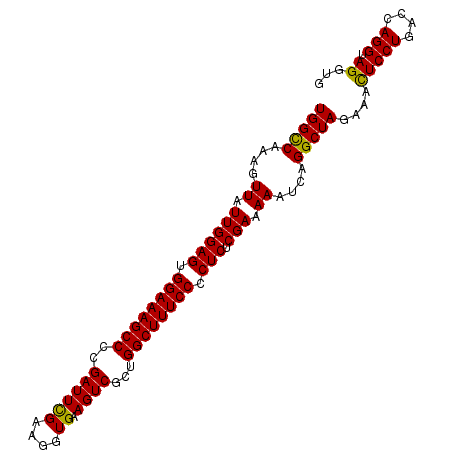
| Location | 5,421,155 – 5,421,254 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -28.27 |
| Energy contribution | -27.65 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.89 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421155 99 + 27905053 UGGCCAAAGUUAUUGGAGUGGAAAGCCCCGAUUCGAAGGUGAAGUCGCUGGCUUUCCCCUCUCGAAAAAUCAGGCUAGAAAUUCCUGACCAGGUAGGUG ..(((.......((((((.((((((((.((((((....)...)))))..)))))))).))).)))....(((((..(....).)))))...)))..... ( -31.80) >DroSec_CAF1 7897 99 + 1 UGGUCAAAGUUAUUGGAGUGGAAAGCCCCGAUUCGAAGGUGAAGUCGCUGGCUUUCCCCUCUCGAAAAAUCAGGCUAAAAAUUCCUGACCAGGUAGGUG (((((.......((((((.((((((((.((((((....)...)))))..)))))))).))).)))......(((.........))))))))........ ( -29.90) >DroEre_CAF1 8367 99 + 1 UGGCCAAAGUUAUUGGAGUGGAAAGCCUCGAUUUGAAGGUGAAGUCUCUGGCUUUCCCCUCUCGAAAAAUCAGGCUAGAAACUCCUGCCCAGGUAGGUG (((((....((.((((((.((((((((..(((((.......)))))...)))))))).))).))).))....)))))....(.(((((....))))).) ( -35.40) >DroYak_CAF1 8608 99 + 1 UGGCCAAAGUUAUUGGAGUGGAAAGCCCCGAUUCGAAGGUGAAGUCUCUGGCUUUCCCCUCUCGACAAAUCAGGCUAGAAACUCCUGACCAGGUAGGUG (((((....((.((((((.((((((((..(((((....)...))))...)))))))).))).))).))....)))))....(((((....))).))... ( -30.30) >consensus UGGCCAAAGUUAUUGGAGUGGAAAGCCCCGAUUCGAAGGUGAAGUCGCUGGCUUUCCCCUCUCGAAAAAUCAGGCUAGAAACUCCUGACCAGGUAGGUG (((((....((.((((((.((((((((..((((((....)).))))...)))))))).))).))).))....)))))....(((((....))).))... (-28.27 = -27.65 + -0.62)


| Location | 5,421,155 – 5,421,254 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 95.62 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -27.86 |
| Energy contribution | -28.05 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421155 99 - 27905053 CACCUACCUGGUCAGGAAUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGCGACUUCACCUUCGAAUCGGGGCUUUCCACUCCAAUAACUUUGGCCA .........(((((((((..((......))..))))(.(((.((((((((..(((.(((......)))))).)))))))).))))........))))). ( -33.60) >DroSec_CAF1 7897 99 - 1 CACCUACCUGGUCAGGAAUUUUUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGCGACUUCACCUUCGAAUCGGGGCUUUCCACUCCAAUAACUUUGACCA .........(((((((((..((......))..))))(.(((.((((((((..(((.(((......)))))).)))))))).))))........))))). ( -31.90) >DroEre_CAF1 8367 99 - 1 CACCUACCUGGGCAGGAGUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGAGACUUCACCUUCAAAUCGAGGCUUUCCACUCCAAUAACUUUGGCCA ..........(((.(((((....((((((((((.....(((((..(((.......)))..)))))))))).)))))....)))))((.....)).))). ( -28.20) >DroYak_CAF1 8608 99 - 1 CACCUACCUGGUCAGGAGUUUCUAGCCUGAUUUGUCGAGAGGGGAAAGCCAGAGACUUCACCUUCGAAUCGGGGCUUUCCACUCCAAUAACUUUGGCCA ......(((....)))........(((.((.((((.(.(((.((((((((...((.(((......)))))..)))))))).)))).)))).)).))).. ( -29.90) >consensus CACCUACCUGGUCAGGAAUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGAGACUUCACCUUCGAAUCGGGGCUUUCCACUCCAAUAACUUUGGCCA .........(((((((((..((......))..))))(.(((.((((((((...((.(((......)))))..)))))))).))))........))))). (-27.86 = -28.05 + 0.19)



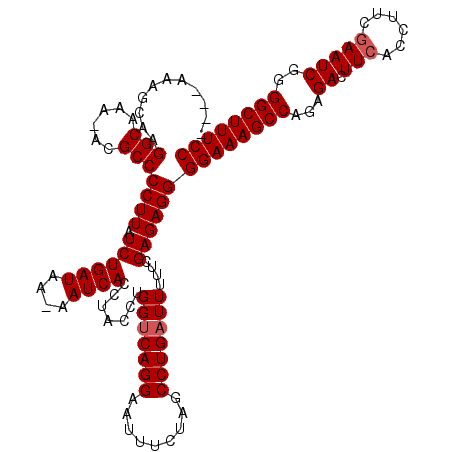
| Location | 5,421,174 – 5,421,293 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.97 |
| Mean single sequence MFE | -34.02 |
| Consensus MFE | -27.33 |
| Energy contribution | -27.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 5421174 119 - 27905053 CUCGAAAGCAAGGCAAA-ACGCCCCUUAUCUGAUAAAAAUCACCUACCUGGUCAGGAAUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGCGACUUCACCUUCGAAUCGGGGCUUUCC ((((((((..((((...-............((((....))))....(((....)))........))))...))))))))...((((((((..(((.(((......)))))).)))))))) ( -36.50) >DroSec_CAF1 7916 118 - 1 CUCGAAAGCAAGGCAAA-ACGCCCCUUAUCUGAUAA-AAUCACCUACCUGGUCAGGAAUUUUUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGCGACUUCACCUUCGAAUCGGGGCUUUCC ((((((((..(((((((-(..((........(((..-.)))(((.....)))..))...)))).))))...))))))))...((((((((..(((.(((......)))))).)))))))) ( -35.60) >DroEre_CAF1 8386 113 - 1 ----AAAGUAAGGCUAA--CGCCCCUUAUCUGAUAA-AAUCACCUACCUGGGCAGGAGUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGAGACUUCACCUUCAAAUCGAGGCUUUCC ----.....((((((..--.((((..((..((((..-.))))..))...)))).(((((((((.((....(((..(....)..))).)).))))))))).............)))))).. ( -30.30) >DroYak_CAF1 8627 115 - 1 ----GAAGCAAGGCAAACACGCCCCUUAUCUGAUAA-AAUCACCUACCUGGUCAGGAGUUUCUAGCCUGAUUUGUCGAGAGGGGAAAGCCAGAGACUUCACCUUCGAAUCGGGGCUUUCC ----.......(((......)))((((.((.(((((-(....((.....))(((((.........)))))))))))))))))((((((((...((.(((......)))))..)))))))) ( -33.70) >consensus ____AAAGCAAGGCAAA_ACGCCCCUUAUCUGAUAA_AAUCACCUACCUGGUCAGGAAUUUCUAGCCUGAUUUUUCGAGAGGGGAAAGCCAGAGACUUCACCUUCGAAUCGGGGCUUUCC ...........(((......)))((((.((((((....)))).......(((((((.........)))))))....))))))((((((((...((.(((......)))))..)))))))) (-27.33 = -27.82 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:22:07 2006